Hairpin structure containing CpG site and monomolecular mechanical method for measuring influence of CpG adjacent sequences on protein dissociation time constant
A hairpin structure and site technology, applied in biochemical equipment and methods, measuring devices, analysis materials, etc., can solve problems such as unclearness and affecting the affinity of protein substrates, and achieve fast collection and improved accuracy Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
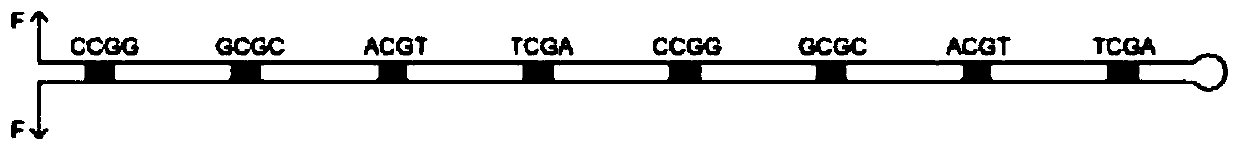
[0092] Embodiment 1: test group comprises the construction of the hairpin structure of CpG site
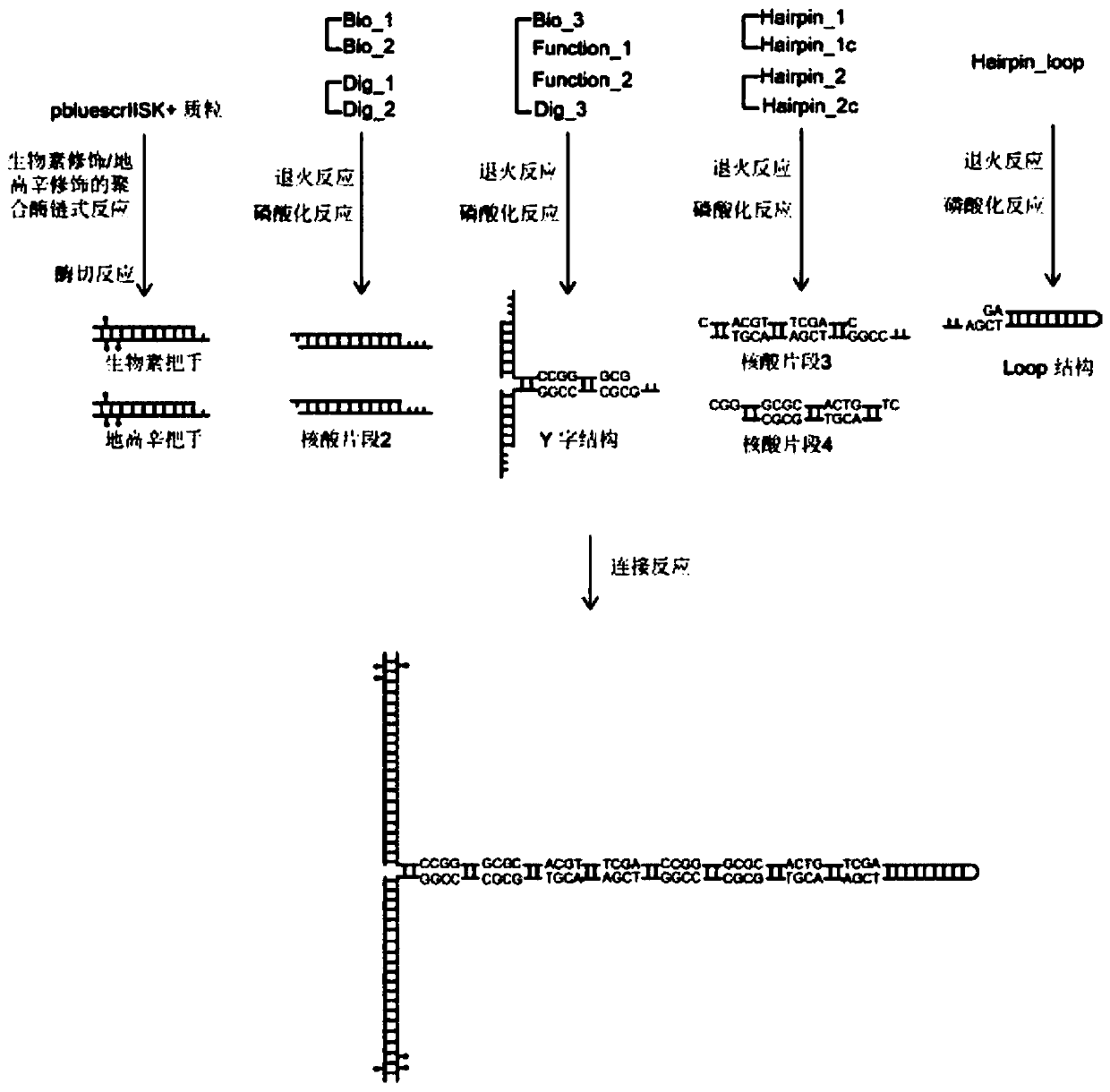
[0093] as per figure 1 The procedure shown produces hairpin structures.
[0094] 1 Preparation of biotin handle and digoxin handle
[0095] 1.1 Polymerase chain reaction of biotin modification / digoxigenin modification
[0096] Perform biotin-modified / digoxigenin-modified PCR on the pbluescrIISK+ plasmid by adding:
[0097] 1 μl of biotin / digoxigenin handle upstream primer at a concentration of 10 μM;
[0098] 1 μl of primers downstream of the biotin / digoxigenin handle with a concentration of 10 μM (both primers were purchased from Jinweizhi Company);
[0099] 1 μl of dATP with a concentration of 10 mM (Cat. No.: 4026Q, Baoriji Biotechnology Co., Ltd.);
[0100] 1 μl of dGTP with a concentration of 10 mM (Cat. No.: 4027Q, Baoriji Biotechnology Co., Ltd.);
[0101] 1 μl of dCTP with a concentration of 10mM (Cat. No.: 4028Q, Baoriyi Biotechnology Co., Ltd.);
[0102] 0.9 μl of...
Embodiment 2
[0181] Embodiment 2: Control group comprises the construction of the hairpin structure of CpG site
[0182] Compared with the construction of the hairpin structure of the CpG site of the test group, the control group needs to additionally synthesize the third nucleic acid fragment of the control group and the fourth nucleic acid fragment of the control group,
[0183] The annealing reaction system of the third nucleic acid fragment of the control group is:
[0184] 8 μl of 50 μM nucleic acid chain whose sequence is shown in SEQ ID NO.18;
[0185] 8 μl of 50 μM nucleic acid chain whose sequence is shown in SEQ ID NO.19;
[0186] 4 μl 5X DNA Annealing Buffer.
[0187] The annealing reaction system of the fourth nucleic acid fragment of the control group is:
[0188] 8 μl of 50 μM nucleic acid chain whose sequence is shown in SEQ ID NO.20;
[0189] 8 μl of 50 μM nucleic acid chain whose sequence is shown in SEQ ID NO.21;
[0190] 4 μl 5X DNA Annealing Buffer.
[0191] The t...
Embodiment 3
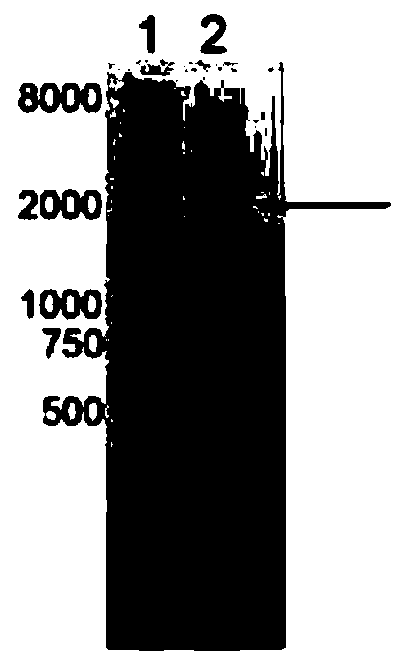
[0205] Example 3: Using single-molecule magnetic tweezers to measure the length change of the hairpin structure under the action of external force
[0206] Making the reaction pool: The reaction pool consists of a slide without holes and a cover glass with holes.
[0207] 1) Punch holes on the cover glass: take a 60mm×24mm glass slide, and use a TC-169 multi-function grinding and engraving machine to punch two holes as the water inlet and water outlet.
[0208] 2) Cleaning of glass slides: Place the coverslip and slide glass vertically in the antibody incubation box, add ultrapure water containing detergent, sonicate in water bath for 30 minutes, rinse with pure water 3 times; add isopropanol, water bath Ultrasonic for 30 minutes, rinse 3 times with pure water; add isopropanol, ultrasonically in water bath for 30 minutes, rinse 3 times with pure water; add pure water, ultrasonically in water bath for 30 minutes, rinse 3 times with pure water; finally add 75% ethanol to soak B...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com