A method for establishing fluorescent nude mouse tumor model based on primary human glioma cells
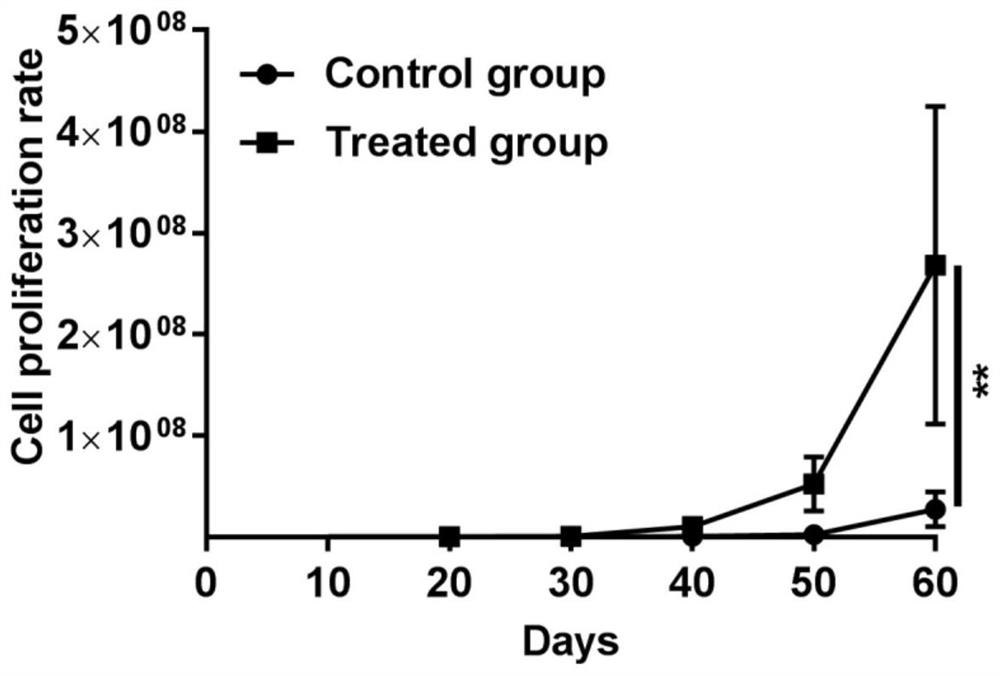
A nude mouse tumor model and human glioma technology, applied to tumor/cancer cells, cells modified by introducing foreign genetic material, animal cells, etc., can solve the problems of background interference, long cycle, complicated operation, etc., and achieve Improve cycle and survival rate, stable survival period and good repeatability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
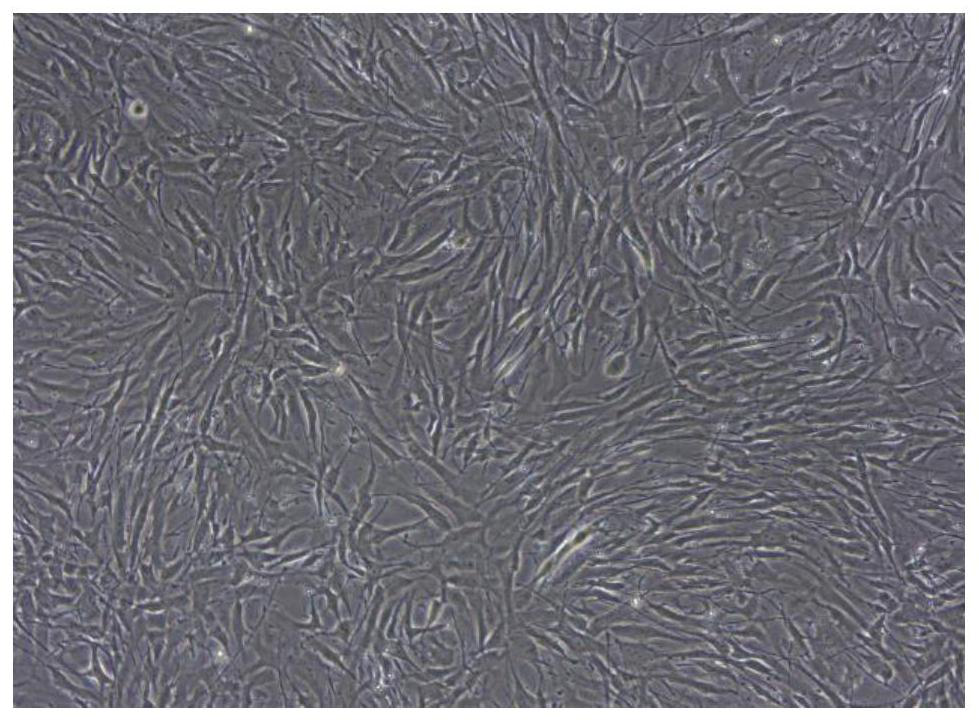
[0038] Example 1 Separation of primary human glioma cells
[0039] (1) Fresh clinical glioma resection specimens were obtained from Wuhan Union Medical College Hospital after passing the hospital ethics committee, agreeing with the patient or the patient's guardian, and signing the informed consent form. The pathological results verified that it was glioblastoma, WHO IV grade.
[0040] (2) Immediately put the resected specimen into pre-cooled sterile tissue preservation solution (containing 1000 U / mL penicillin, 1000 μg / mL streptomycin sulfate, 2.5 μg / mL amphotericin and 50 μg / mL gentamicin) Into the collection tube, and immediately put into a 4 ℃ sample transport box, transported to the laboratory within 4 hours for cell separation.
[0041] (3) Separation of primary cells: Obtain the tissue in a biological safety cabinet, rinse it once with absolute ethanol, and rinse it twice with 1×PBS (pH7.2-7.4), remove blood vessels, For fat and necrotic tissue, use dissecting scissors...
Embodiment 2
[0042] Example 2 Subculture of primary human glioma cells and optimization of subculture conditions
[0043] The isolated primary cells were divided into four groups, which were subcultured in different modified DMEM / F12 complete medium (medium A-D) conditions;
[0044] (1) Resuspend the cell pellet with modified DMEM / F12 complete medium, and store at 37°C, 5% CO 2 Cultivate under certain conditions until the cell abundance in the T25 culture flask reaches 80%, rinse the cells twice with 1×PBS (pH7.2-7.4), add 1 mL of 0.05% trypsin-EDTA to digest the monolayer cells 2- 3min.
[0045] (2) Add 2 mL of improved DMEM / F12 complete medium to stop the digestion; centrifuge at 1000 rpm for 4 minutes, remove the supernatant, collect the cell suspension, resuspend with 1 mL of improved DMEM / F12 complete medium and supplement the medium, according to the ratio of 1 to 2 , placed in a T25 culture bottle for culture, and after 20 generations of continuous passage, it is ready for use.
...
Embodiment 3
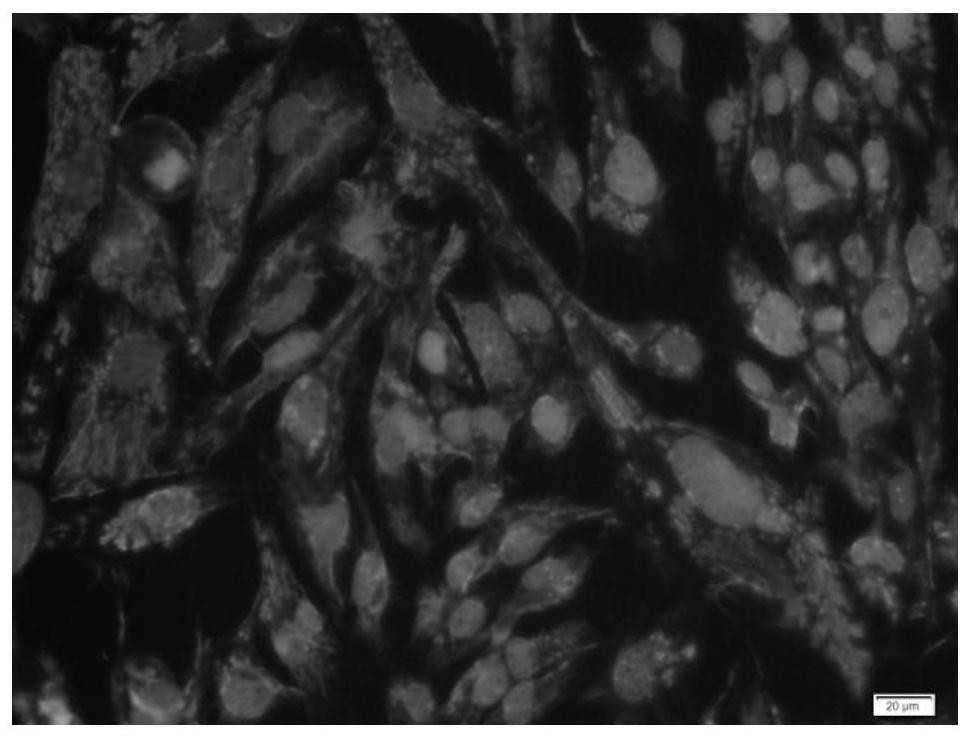
[0054] Example 3 Identification of Specific Marker Expression in Human Brain Glioma Primary Cells
[0055] (1) In the culture plate, soak the slide with the cells climbed in PBS for 3 times, each time for 3 minutes;
[0056] (2) Fix slides with 4% paraformaldehyde for 15 minutes, soak and wash slides with PBS 3 times, 3 minutes each time;
[0057] (3) Permeate with 0.5% TritonX-100 (prepared in PBS) for 20 minutes at room temperature;
[0058] (4) PBS soaked slides 3 times, 3 minutes each time, blotted the PBS with absorbent paper, added normal goat serum to the slides, and sealed at room temperature for 30 minutes;
[0059] (5) Absorb the blocking solution with absorbent paper, without washing, add a sufficient amount of diluted primary antibody to each slide and put it in a wet box, and incubate overnight at 4°C;
[0060] (6) The next day, soak the slides with PBST for 3 times, each time for 3 minutes, absorb the excess liquid on the slides with absorbent paper, add the di...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com