Primer group and kit for single-tube detection of human spinal muscular atrophy
A spinal muscular atrophy, human detection technology, applied in recombinant DNA technology, microbial determination/examination, biochemical equipment and methods, etc. Application requirements, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0152] The selection of the primer design position of embodiment 1 highly homologous SMN1 and SMN2
[0153] Amplification primers include universal sequences (or specific sequences) and recognition sequences, among which the universal primers are used to realize the simultaneous amplification of different amplicons, and can bind and extend the amplification products of different recognition sequences, finally in the amplification program The upper Tm value is higher and the second cycle amplification; specific sequence, similar to the tag sequence on the next generation sequencing method, the designed primers for different targets include different nucleotide lengths, which are used to distinguish different SMN amplifications in length sub-length; recognition sequence, the recognition sequence on different primers can specifically bind to exon 7 of SMN1 and SMN2, exon 4 and exon 5 of NIAP gene, and exon 10 of GTF2H2 gene respectively , and extend and amplify. The following ma...
Embodiment 2
[0161] Example 2 Amplification blocking primer design of SNP site difference and amplification blocking primer design for copy number quantification of SMN1 / 2 and "2+0" carrier
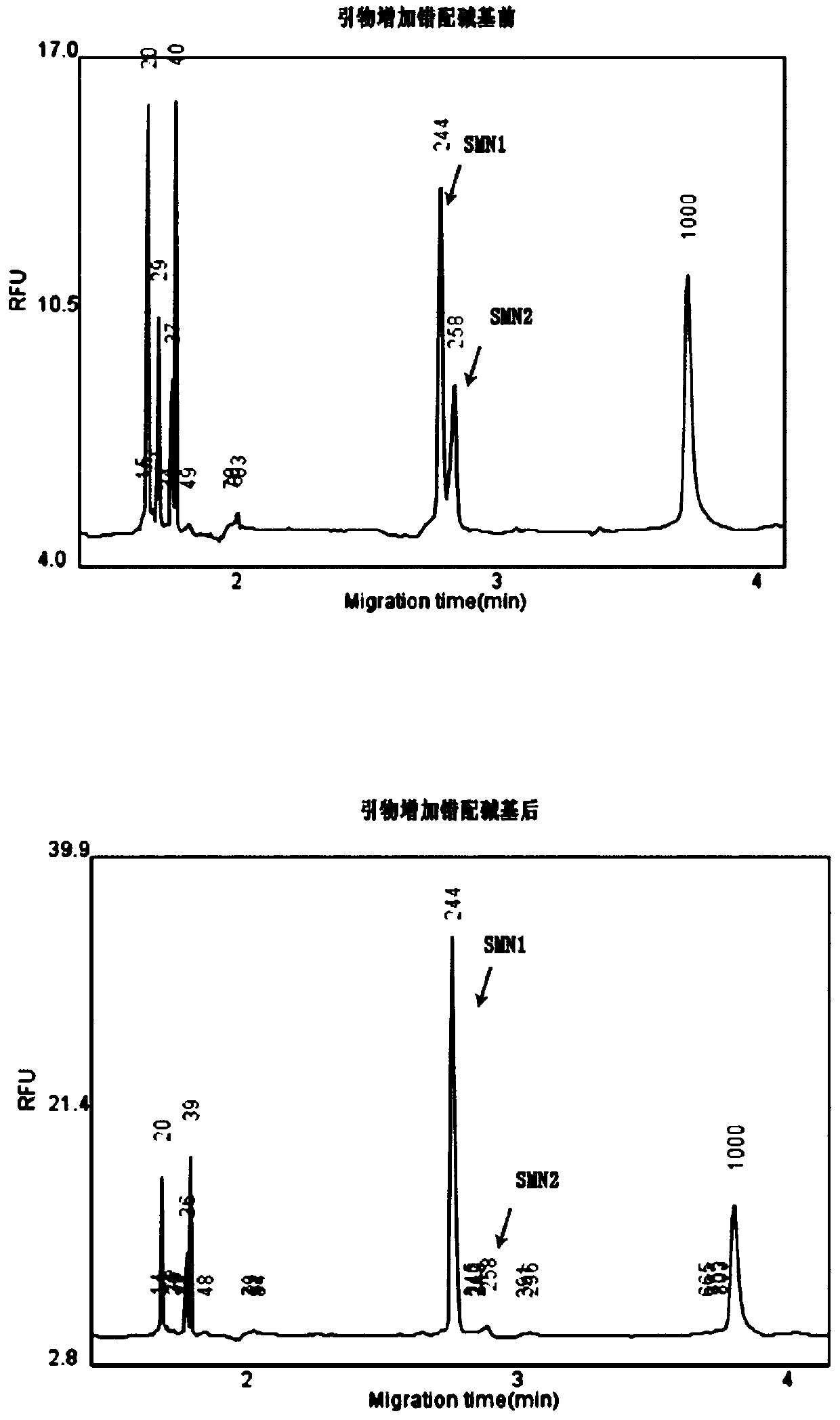
[0162] For the difference base between SMN1 and SMN2, the SNP mutation site on the SMN1 gene, etc., the 3' end of the sequence adopts the amplification block design, which is different from the general amplification block design, which can only match the template at the 3' end, while the There is a mismatch with the homologous template, and the present invention introduces an additional 1 to 3 mismatched bases in the 2 to 5 bases at the 3' end. Based on the strength of base matching, while ensuring the quality of primers (try to avoid primer dimers, mismatches, hairpin structures, etc.), additional mismatched bases are introduced.
[0163] Primer design for different positions:
[0164] (1) Design primers for the differential bases in the 7th intron (+100A) of SMN1, except that the 3' end of the prim...
Embodiment 3
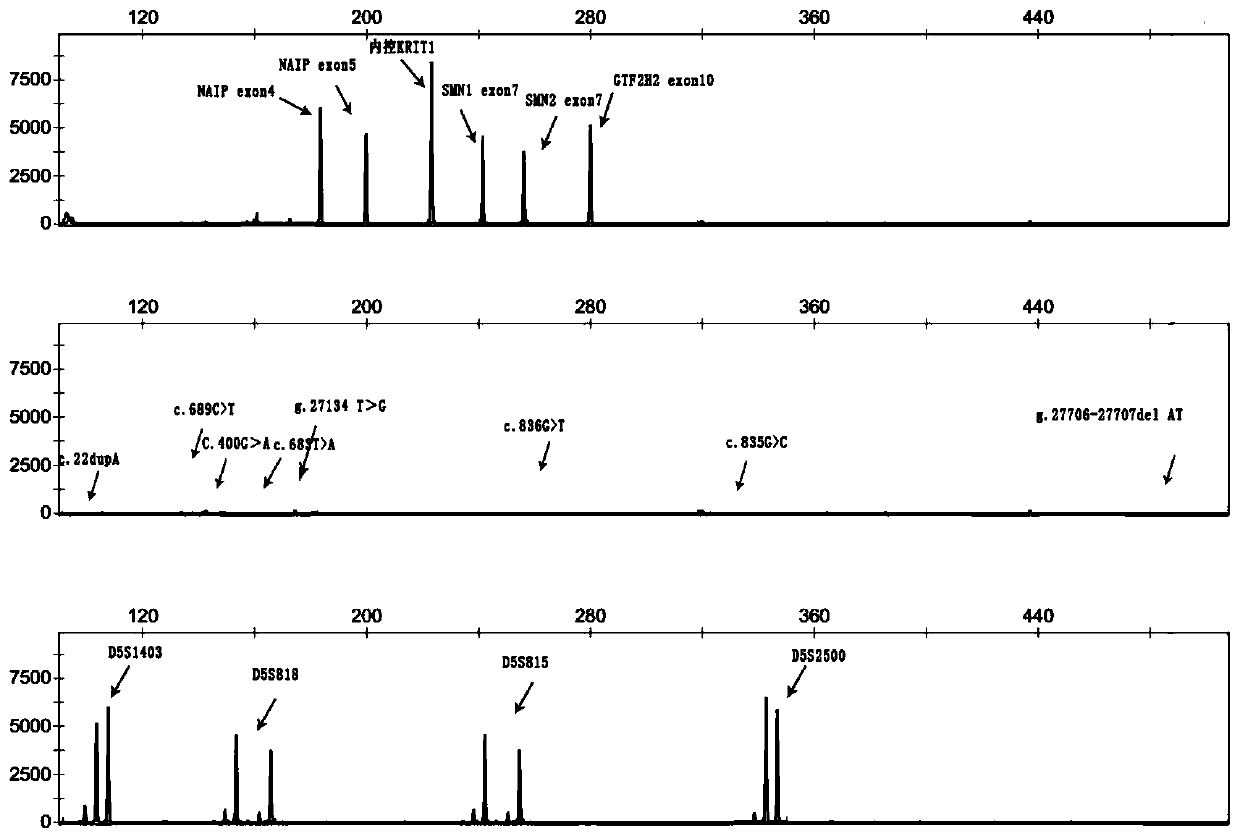
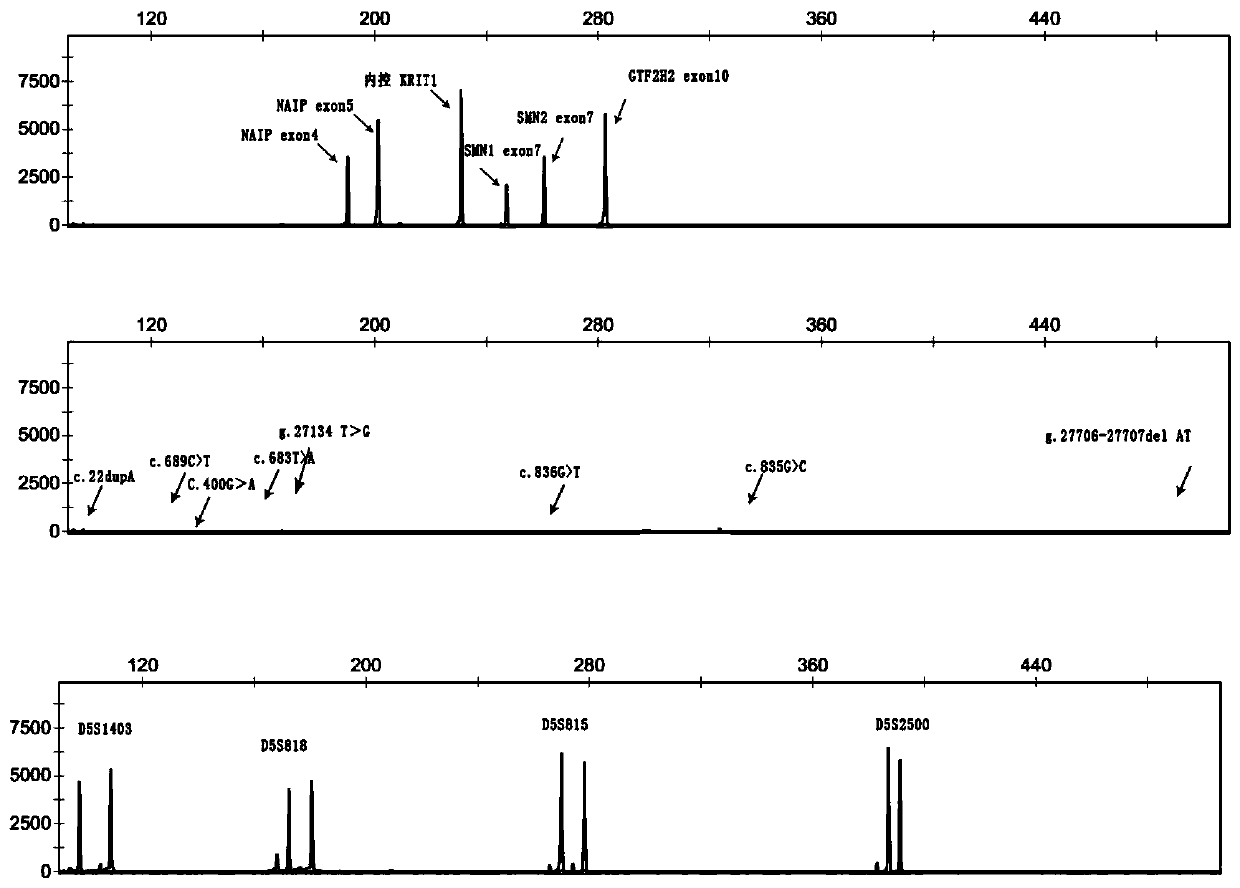
[0174] Example 3 Single-tube human spinal muscular atrophy-related gene copy number and point mutation detection, carrier typing kit
[0175] 1. Composition
[0176] The kits of the present invention are designed according to 50 people, including: 5 × PCR reaction mixture, 1 tube (275 μL / tube); primer mixture, 1 tube (138 μL / tube); PCR enzyme mixture, 1 tube (28 μL / tube); tube); SMN calibrator, 1 tube (275 μL / tube); SMN quality control product, 1 tube (275 μL / tube); diluent, 1 tube (1000 μL / tube).
[0177] Among them, the 5×PCR reaction mixture includes Tris-Hcl buffer 175mmol / L, potassium chloride 200mmol / L, ammonium sulfate 83mmol / L, calf serum albumin 500μg / mL, gelatin 0.05%, Tween-200.5%, di Thiothreitol 25mmol / L, dATP 1000nM, dGTP 1000nM, dCTP 1000nM, dTTP 500nM, dUTP 1000nM.
[0178] This kit designs primers based on the base difference between Exon 7 and Intron 7, and the PCR product is generally "Exon 7 of SMN1 + Intron 7 partial sequence" or "Exon 7 of SMN2 No. exo...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com