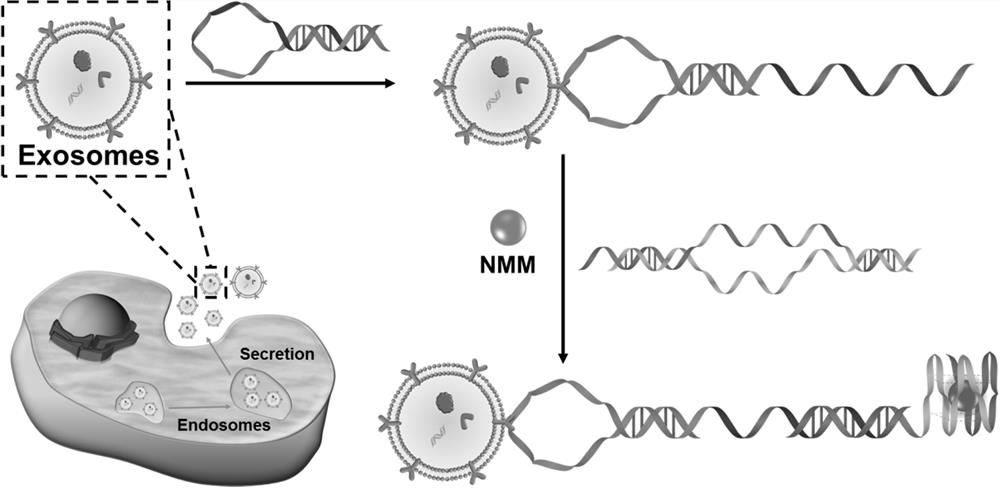
A self-assembled probe based on conformational change and its label-free detection method for exosomes
A technology of configuration change and detection method, applied in the field of molecular biology, can solve the problems of affecting the efficiency of identification, background interference and so on
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036] In this embodiment, the self-assembly probe based on configuration change is used for the label-free detection method of exosomes, and the steps are as follows:
[0037] (1) Cell culture: CCRF-CEM, Ramos, Hela, B16F1 and HL-7702 cells were cultured in RPMI 1640 medium supplemented with 10% exosomes (fetal bovine serum removed) and 100IU / mL penicillin or streptomycin cultured at 37 °C with 5% wt / vol CO 2 Incubate in a humidified incubator for 48 hours, and collect the cell supernatant when the cells grow to 80-90%;
[0038] (2) Isolation of exosomes: The cell supernatant obtained in step (1) was centrifuged at 3,000g for 30 minutes at 4°C, and filtered through a syringe-driven filter unit to remove intact cells and cell debris, and the collected filter and concentrated using 100 KDa MWCO at 5,000 g for 30 min at 4 °C, then the filtered medium was ultracentrifuged at 160,000 g for 2 h at 4 °C, the supernatant was discarded, and the exosome pellet was resuspended in phospha...
Embodiment 2
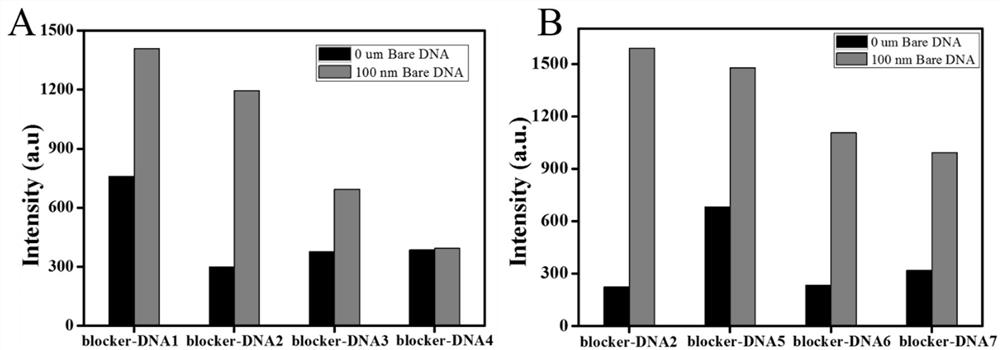
[0042] In this example, in order to further study the hybridization and binding ability of G4-DNA and different blocker-DNAs, eight groups of Blocker DNA sequences were designed, such as SEQ ID NO.3, SEQ ID NO.4, SEQ ID NO.5, SEQ ID NO. .6, SEQ ID NO.7, SEQ ID NO.8, SEQ ID NO.9 and SEQ ID NO.10; refer to the procedure of Example 1 for fluorescence detection. First, a Bare DNA strand was designed, which contained a complete base of the priming region to simulate the probe recognizing exosomes, which could be used to trigger a strand displacement reaction to release G4. Test results such as figure 2 shown, by figure 2 It can be seen that by using BlockerDNA-2, when Bare DNA is present, the maximum fluorescence enhancement can be obtained, which is significantly higher than that of blocker DNA-1, blockerDNA-3, blocker DNA-4, Blocker DNA-5, Blocker DNA-6 and Blocker DNA- 7. Therefore, blocker DNA-2 was selected as the best blocker DNA for subsequent experiments and named R-DN...
Embodiment 3
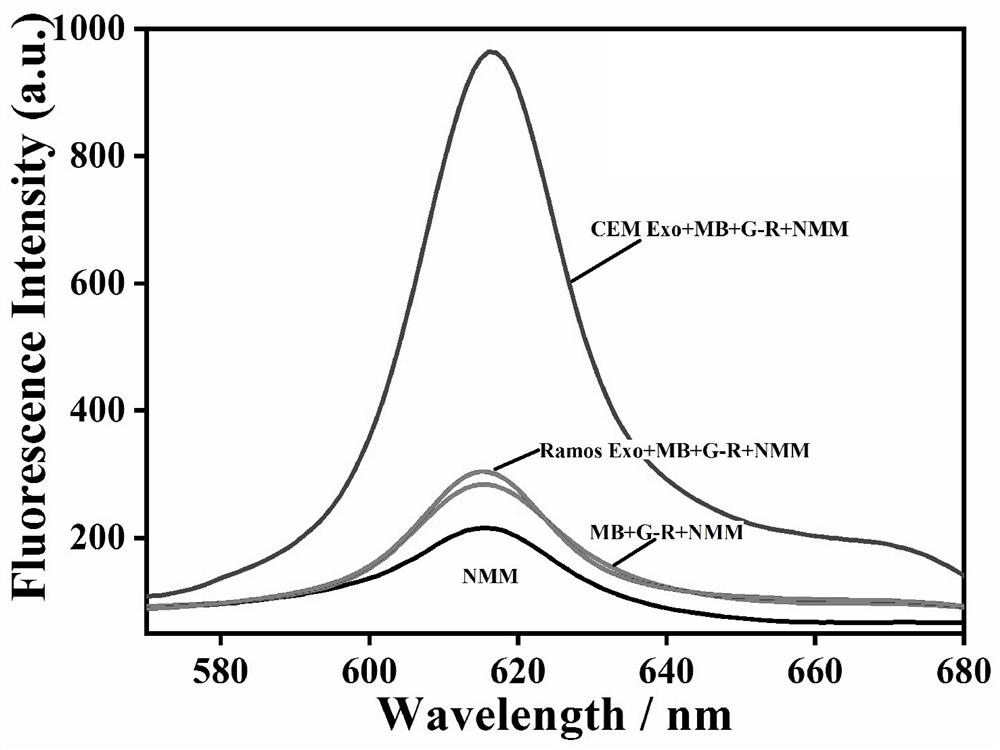
[0044] In this example, in order to prove the feasibility of the experimental principle, the fluorescence intensity of the system under different conditions was studied. The test results are shown in image 3 shown, by image 3 It can be seen that in the absence of target exosomes (red curve) and control exosomes (blue curve), there is no obvious fluorescence signal, indicating that the present invention has a lower fluorescence background. After the target exosomes were introduced into the above mixed solution, an obvious fluorescence enhancement could be observed at around 615 nm (green curve), which indicated that the G-rich sequence was exposed and folded into a quadruple structure. These results suggest that this experimental platform can be used for the specific detection of leukemia-related exosomes.
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com