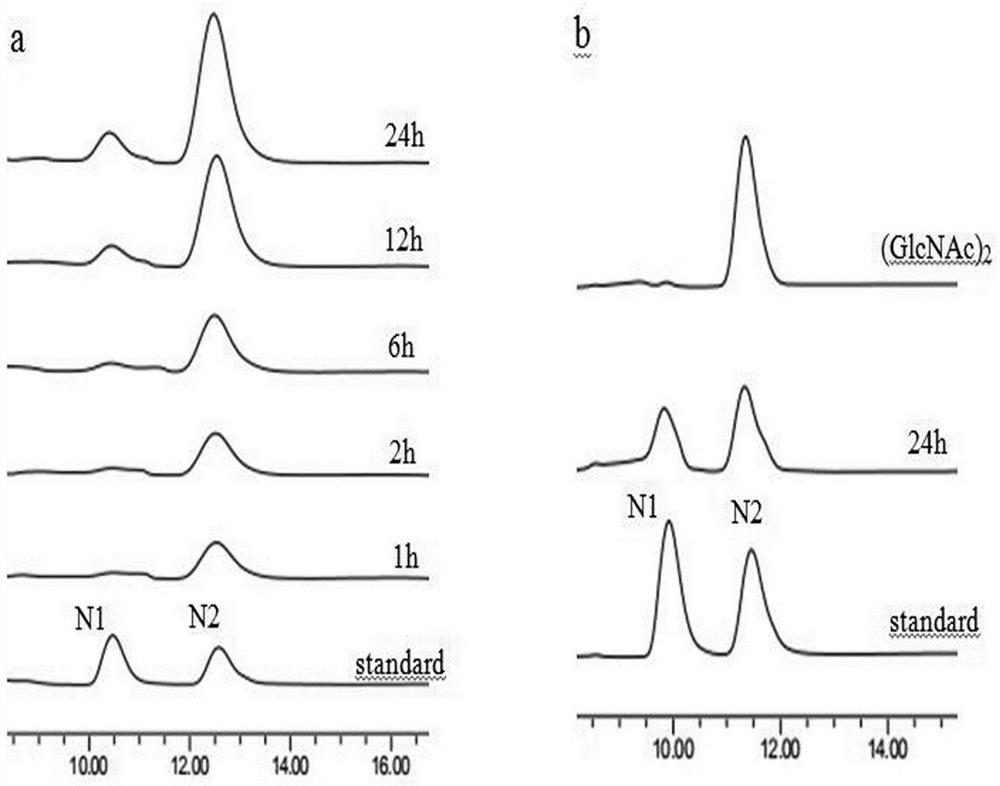
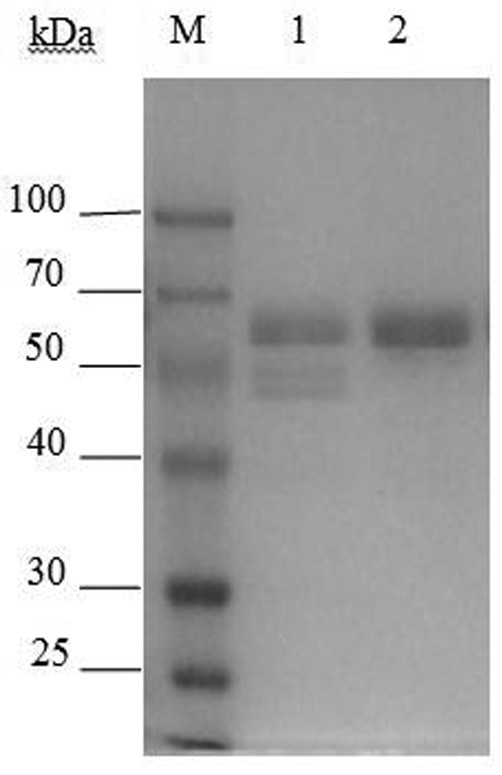
A chitinase and its gene for preparation of chitooligosaccharides
A chitinase gene and chitinase technology are applied in the fields of glycosylase, genetic engineering, and plant gene improvement, which can solve the problems of enzyme inactivation, long growth cycle, and reduction of enzyme activity, and achieve high activity and stability, maintaining activity and stability, and efficient degradation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0040] Example 1 Chitinase gene chi1602 the acquisition
[0041] (1) Microbulbifer sp . Separation and extraction of genomic DNA:
[0042] Take 1.5 mL bacterial culture Microbulbifer sp . (purchased from China Marine Microorganism Culture Collection Management Center) in a sterilized Ep tube, centrifuged at 12000 rpm for 1 min, discarded the supernatant, and collected the bacteria.
[0043] Add 400 μL of lysate solution (40 mM Tris-acetic acid, 20 mM sodium acetate, 1 mM EDTA, 1% SDS, pH 7.8), mix well, and place in a 37°C water bath for 1 h.
[0044] Then add 200 μL of 15 mol / L sodium chloride solution, mix well and centrifuge at 13000 rpm for 15 min.
[0045] Take the supernatant, extract twice with phenol and once with chloroform.
[0046] Add twice the volume of absolute ethanol and 1 / 10 volume of potassium acetate (3 M, pH 8.0), store at -20°C for 1 h, centrifuge at 13,000 rpm for 15 min, discard the supernatant, and wash the precipitate twice with 70% ethanol;...
Embodiment 2
[0057] Example 2 Expression and amplification of chitinase-encoding gene chi1602 in Escherichia coli
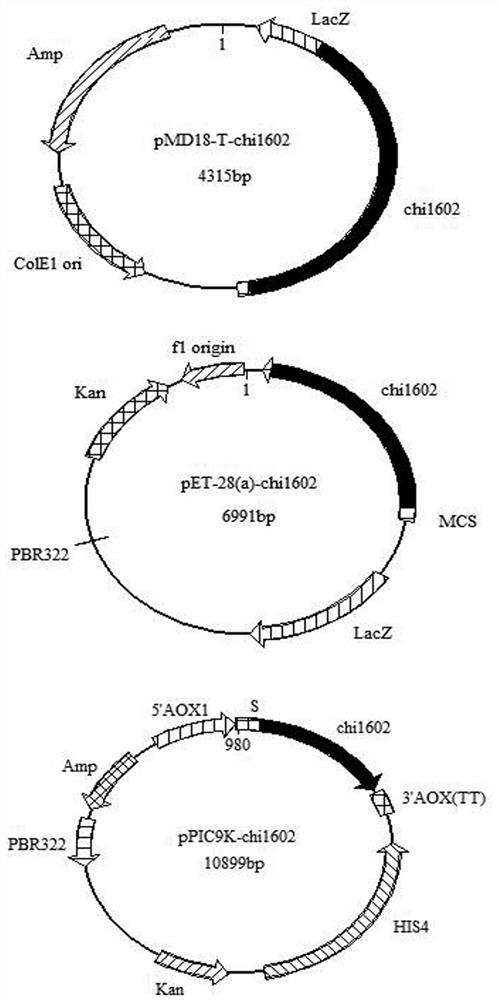
[0058] The result obtained in embodiment 1-(4) Sec I and not I double-enzyme-digested target gene, and Sec I and not The pET-28a (+) plasmid of I double digestion is connected, obtains recombinant plasmid pET-28 (a)- chi1602 (Such as figure 1 shown).
[0059] Take 10 uL of the constructed plasmid DNA and add it to 100 uL of prepared Escherichia coli BL21(DE3) competent cells, shake well and place on ice, ice bath for 30 min; place in a 42°C water bath for 45 s heat shock; put Quickly move the centrifuge tube to ice-water mixture for 2 min; add 800 uL LB liquid medium to each tube, and recover on a shaker at 37°C for 1 h (80 rpm to 200 rpm); centrifuge at 4000 rpm for 5 min, and discard 800 uL Clear, and mix the rest; smear on a plate (LB-agar plate, containing 100 ug / mLAmp), culture overnight at 37°C, pick the recombinants into LB liquid medium, wait until the bact...
Embodiment 3
[0061] Example 3 Construction of Yeast Recombinant Expression Vector
[0062] The result obtained in embodiment 1-(4) SnaB I and Avr ⅡThe target gene of double enzyme digestion, and after SnaB I and Avr Ⅱ The double-digested pPIC9k plasmid was ligated to obtain the recombinant plasmid pPIC9k- chi1602 (Such as figure 1 shown).
[0063] Transfer the recombinant plasmid into E. coliTop10 competent cells were spread on the LB screening plate containing 100 mg / mL Amp and cultured overnight, and the recombinants were picked and identified by bacterial liquid PCR. If they were positive recombinants, they were sent for sequencing, and the sequencing results further proved the sequence of the target gene and the correctness of the insertion site. Select the bacterial solution with the correct inserted gene, follow the extraction steps of the OMEGA PlasmidMini Kit, extract the recombinant plasmid, and store it at -20°C for later use.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com