PCR-CRISPR detection method for targeting HBV drug-resistant mutant gene
A PCR-CRISPR and drug-resistant gene technology, applied in the field of molecular biology, can solve the problems of high cost, easy pollution, unsuitable for clinical detection, etc., and achieve the effect of strong fluorescence detection signal and simple method
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0056] Embodiment 1: be used for the design and preparation of crRNA of the present invention and PCR primer
[0057](1) Design of YVDD and YIDD drug-resistant mutant crRNA
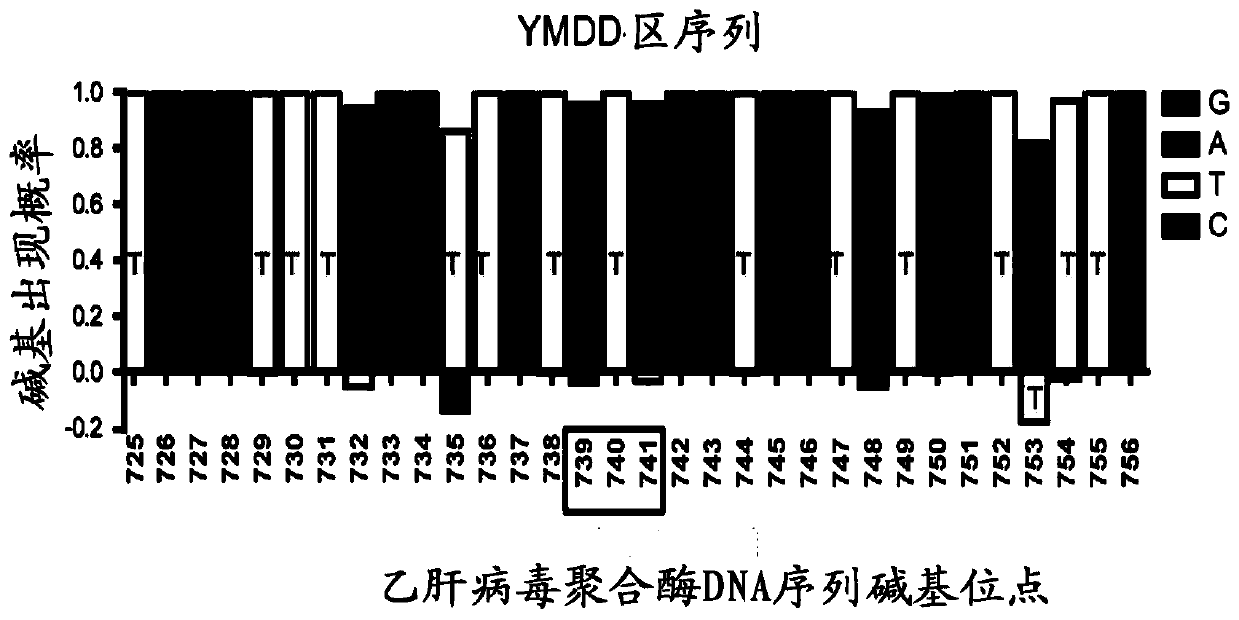
[0058] A total of 32bp (725-756bp) nucleotides in the upstream and downstream of the YMDD region were analyzed for sequence conservation (see figure 2 ), for the less conserved sites, select the base with the highest probability of each site to design crRNA. figure 2 The middle box marks the rt204 site, and the analysis results show that, except for the three sites nt732, nt735 and nt753, the sequences of other sites are relatively conserved. According to the conservation characteristics of this partial sequence, we designed corresponding crDNA templates and PCR primers (see Table 2) for preparing crRNA for mutation detection. The DNA sequence was synthesized by Beijing Tianyi Huiyuan Company.
[0059] Table 2. crDNA templates and PCR primers used to prepare crRNA for mutation detection
[0060] ...
Embodiment 2
[0084] Embodiment 2.Specific detection of YVDD and YIDD drug-resistant mutation crRNA
[0085] The target sequences of the wild strain of hepatitis B virus and the two drug-resistant mutant strains were transcribed into corresponding ssRNA, and the specificity of the three sequences were detected by using the above-mentioned two drug-resistant mutant crRNAs.
[0086] (1) Detection of mutant crRNA to wild-type and mutant sequences
[0087] Using the wild-type and two drug-resistant mutant plasmids constructed above and the designed primers HBV-F and HBV-R, amplification, transcription, and purification of ssRNA were carried out. The reaction system and conditions were the same as before. Two kinds of crRNA were used to detect ssRNA of three sequences respectively, and the detection system was prepared as follows:
[0088] Table 6. ssRNA Target Cutting System
[0089]
[0090]
[0091] Then, at 37°C, the fluorescent signal of the FAM channel detection system of the fluor...
Embodiment 3
[0103] The sensitivity detection of embodiment 3.YMDD drug resistance mutation
[0104] (1) Agarose gel electrophoresis detection
[0105] YVDD, YIDD mutant plasmids and YMDD wild-type plasmid standard products were serially diluted and used as templates for PCR amplification, followed by 1.5% agarose gel electrophoresis detection, and the electrophoresis results were as follows: Figure 8 shown. Figure 8 middle. A. The electrophoresis results of the HBV WT wild-type plasmid gradient dilution PCR, B. the electrophoresis results of the HBV YIDD mutant plasmid gradient dilution PCR, and C. the electrophoresis results of the HBV YVDD mutant plasmid gradient dilution PCR. The negative control (NC) corresponding to the three groups of templates had no bands, indicating that there was no pollution during the amplification process; the results showed that a single band with the same size as the target band (304bp) appeared between 250-500bp; at the same time, the three Group temp...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com