One-step nucleic acid test method based on CRISPR/Cas and isothermal amplification and kit
A detection method, constant temperature amplification technology, applied in the direction of biochemical equipment and methods, microbial measurement/inspection, etc., can solve the problems of long detection time, expensive price, complicated operation, etc., to ensure accuracy, convenient and rapid detection , The effect of improving the ease of operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0041] In the embodiment of the present invention, based on CRISPR / Cas12a technology, on-site rapid lateral flow immunochromatography test paper detects nucleic acid. This method is used for non-diagnostic or therapeutic purposes, and includes the following steps:
[0042] 1) Design and transcribe crRNA: find the crRNA target sequence (such as SEQ ID NO: 1) that conforms to the TTTN 18N principle in the target nucleic acid sequence to be tested, and determine the crRNA sequence from it. RPA upstream and downstream amplification primers (such as SEQ ID NO: 2-3) were designed within 200 bp of both ends of the target sequence.
[0043] The method of crRNA in vitro transcription is as follows: react at 37°C for 16 hours in the following system: add Nuclease-free water to 20 μL, NTP 1.5 μL each, 10×reaction buffer 1.5 μL, Template DNA 1 μg, T7RNA Polymerase Mix 1.5 μL.
[0044] CrRNA purification method: Mix 20 μL of in vitro transcription product with 160 μL nuclease-free water an...
Embodiment 2
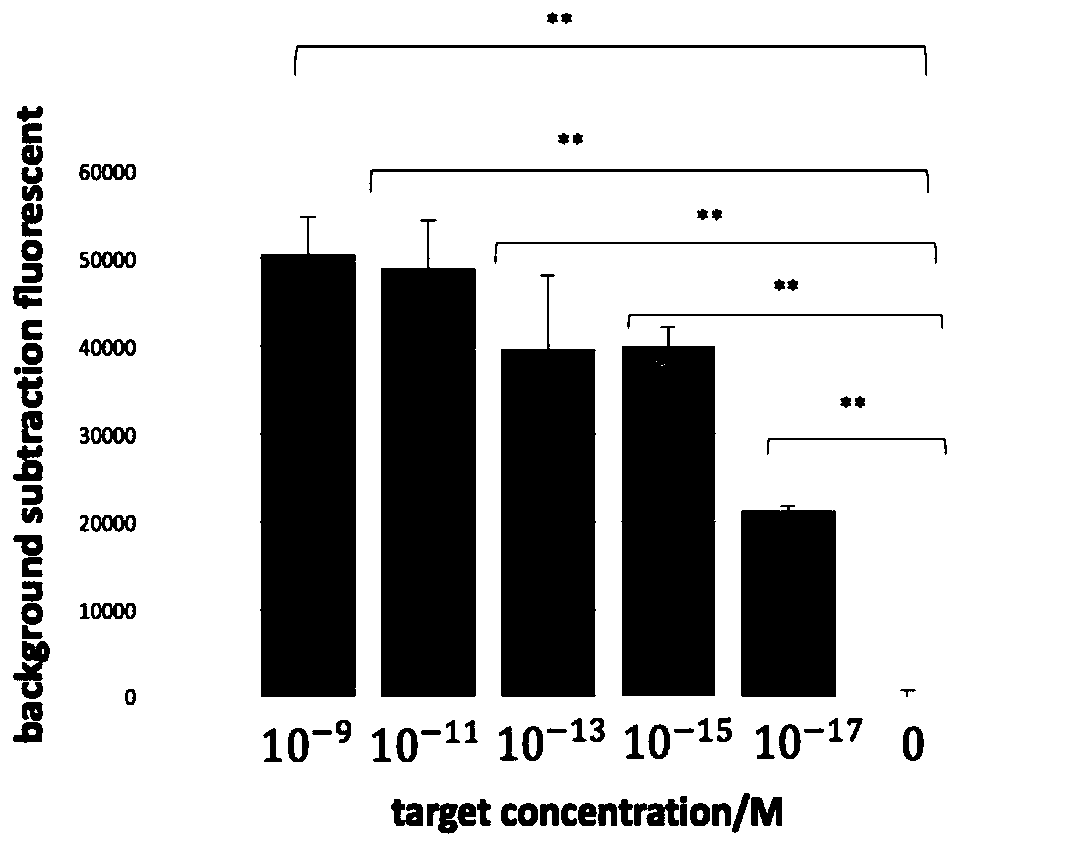
[0052] In the embodiment of the present invention, based on CRISPR / Cas12a technology, on-site rapid fluorescent detection of nucleic acid, the method is used for non-diagnostic or therapeutic purposes, including the following steps:
[0053] 1) Design and transcribe crRNA: find the crRNA target sequence (such as SEQ ID NO: 1) that conforms to the TTTN 18N principle in the target nucleic acid sequence to be tested, and determine the crRNA sequence from it. RPA upstream and downstream amplification primers (such as SEQ ID NO: 2-3) were designed within 200 bp of both ends of the target sequence.
[0054] The method of in vitro transcription of crRNA is as follows: react at 37°C for 16 hours in the following system: add Nuclease-free water to 20 μL, NTP 1.5 μL each, 10×reaction buffer 1.5 μL, Template DNA 1 μg, T7 RNA Polymerase Mix 1.5 μL.
[0055] CrRNA purification method: Mix 20 μL of in vitro transcription product with 160 μL nuclease-free water and 20 μL 3M ammonium acetate,...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com