Method for detecting methylation modification of adenine N6 or N1 locus in nucleic acid by means of dUTP or dTTP
A methyladenine and nucleic acid technology, applied in the field of identification and detection of N6-methyladenine or N1-methyladenine in the genome, to achieve the effect of mild experimental conditions
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
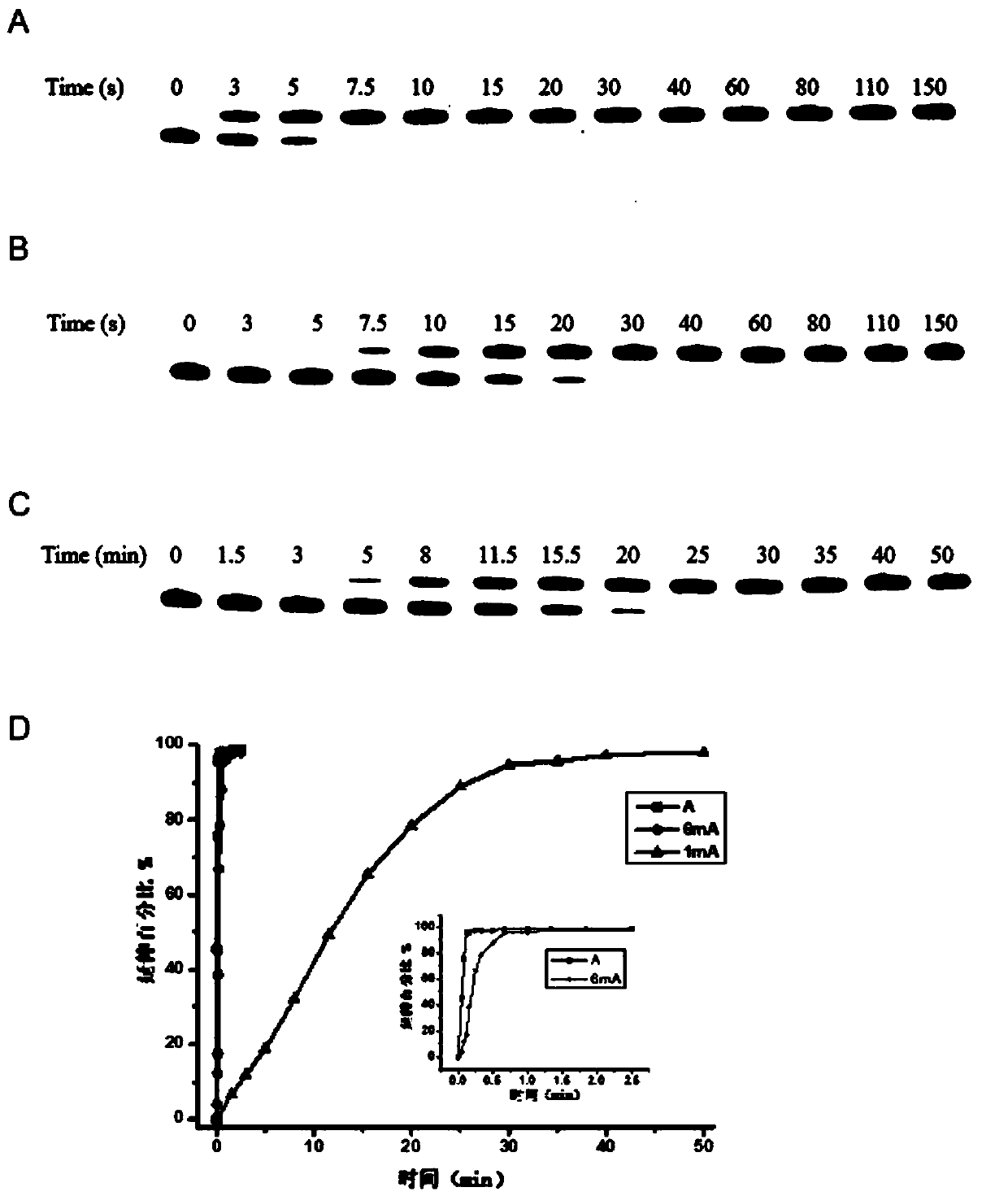
[0049] The method of the invention is applied to sequence detection under 25 μM dUTP. The specific steps are as follows:
[0050] 1.6-carboxyfluorescein (6-carboxy-fluorescein, FAM) has a maximum excitation wavelength of 495nm and a maximum absorption wavelength of 521nm, which can be used to label the 5' end of the DNA primer to be tested. Under the excitation of ultraviolet light in a gel imager, No re-staining is required for imaging;
[0051] 2. Prepare 1 μL of 1×ThermoPol buffer at pH 8.8 at 25° C. according to the following formula: 20 mM Tris-HCl, 10 mM ammonium sulfate, 10 mM potassium chloride, and 2 mM magnesium sulfate. Add 0.1% polyethylene glycol octylphenyl ether, 0.2 μL double-stranded DNA with a final concentration of 2 μM (containing adenine at the same position, N 6 -Methyladenine, N 1-methyladenine) DNA-17mer-A1, DNA-17mer-6mA1 and DNA-17mer-1mA1, 25 μM dUTP, 1 μL Bst DNA polymerase, and primer Primer, the concentration ratio of primer and double-stranded...
Embodiment 2
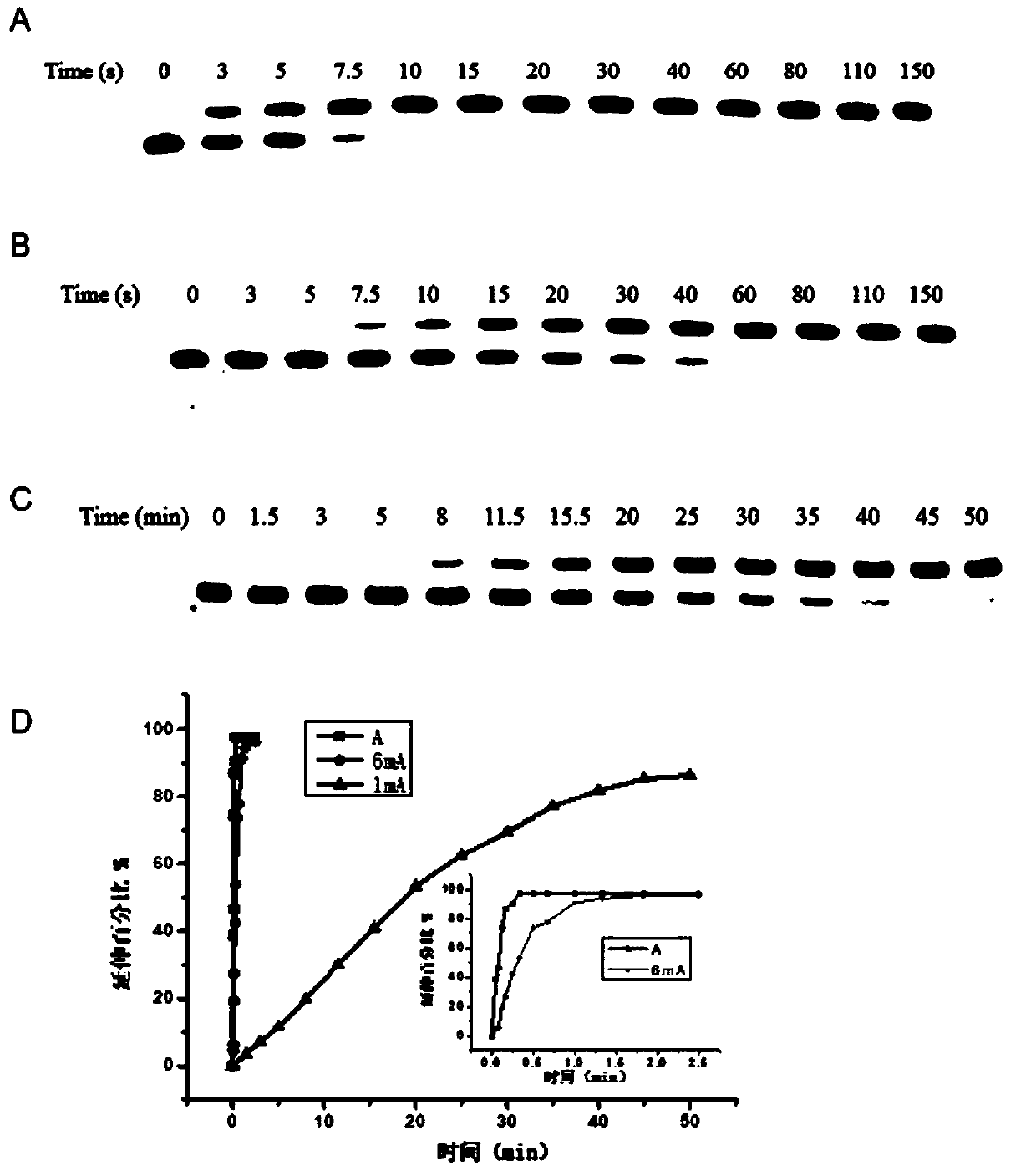
[0056] The method of the present invention is applied to sequence detection under 6.25 μM dTTP, and the specific steps are as follows:
[0057] 1. Prepare 1 μL of 1×ThermoPol buffer at pH 8.8 at 25°C: 20 mM Tris-HCl, 10 mM ammonium sulfate, 10 mM potassium chloride, and 2 mM magnesium sulfate. Add 0.1% polyethylene glycol octylphenyl ether, 0.2 μL double-stranded DNA with a final concentration of 2 μM (containing adenine at the same position, N 6 -Methyladenine, N 1 -Methyladenine) DNA-17mer-A1, DNA-17mer-6mA1 and DNA-17mer-1mA1, 6.25 μM dTTP, 1 μL Bst DNA polymerase, and primer Primer, the concentration ratio of primer and double-stranded DNA is 3:5 , and adding ddH 2 O make up the volume to 10 μL;
[0058] 2. Incubate at a controlled constant temperature of 63° C. for 5 minutes, take samples for analysis at different intervals, and add 45 μL of stop buffer (95% formamide, 25 mM EDTA, pH 8.0) to stop the reaction during analysis. Immediately after termination, heat to 90°...
Embodiment 3
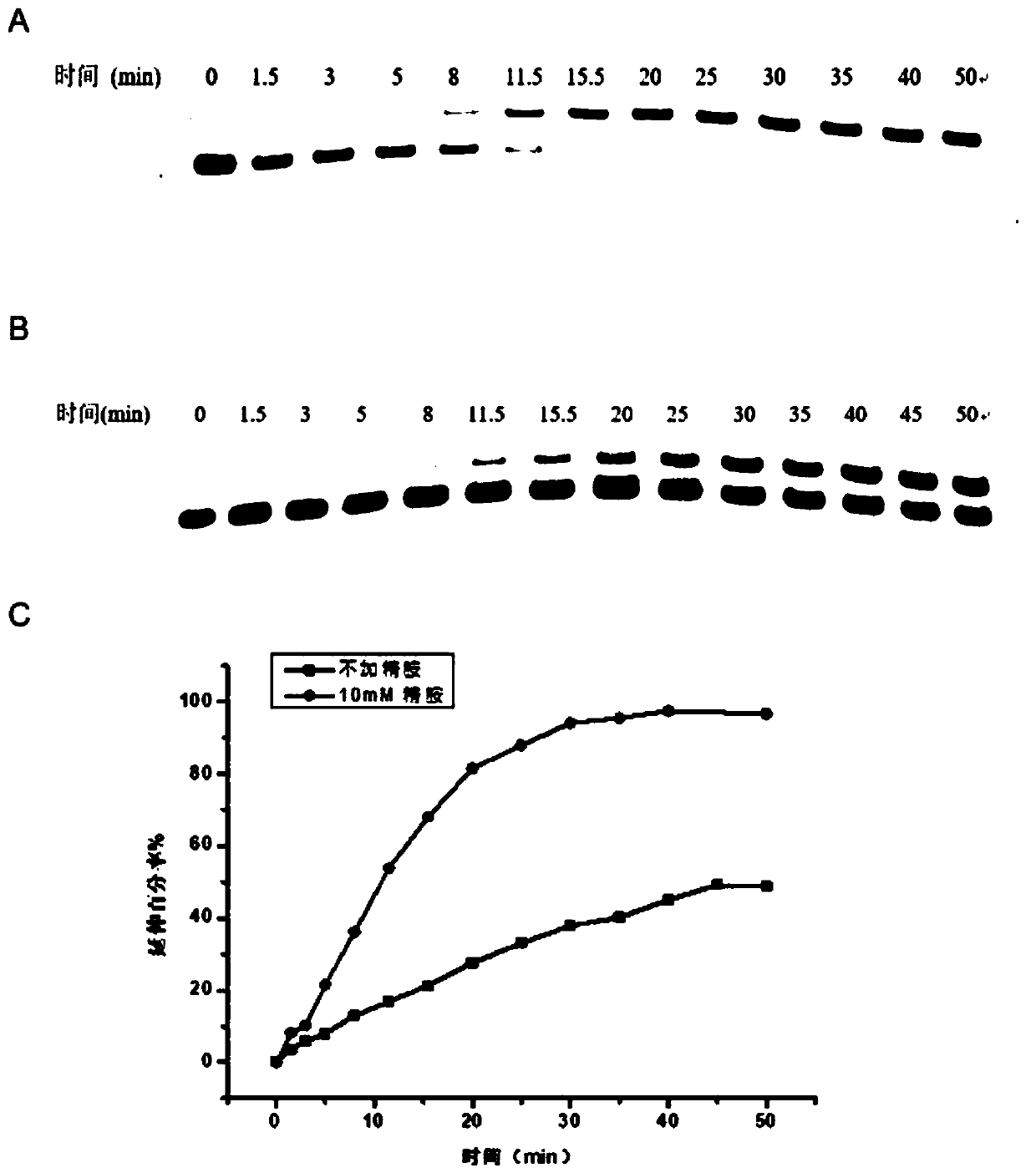
[0062] The method of the present invention is used to explore whether spermine exists to contain N 1 -The detection impact of the sequence of the methyladenine site, the specific steps are as follows:
[0063] 1. Prepare two groups of 1 μL 1×ThermoPol buffer solution at pH 8.8 at 25° C. according to the same recipe as in Examples 1 and 2. Add 0.1% polyethylene glycol octylphenyl ether, 0.2 μL double-stranded DNA DNA-17mer-1mA1 with a final concentration of 2 μM, dTTP with a final concentration of 6.25 μM, 1 μL Bst DNA polymerase, and primer Primer, primer and double-stranded The concentration ratio of DNA was 3:5, one group was added with a final concentration of 100 μM spermine, the other group was used as a blank control, and ddH was added to both systems 2 O make up the volume to 10 μL.
[0064] 2. Incubate at a controlled constant temperature of 63° C. for 10 min, and add 45 μL of stop buffer (95% formamide, 25 mM EDTA, pH 8.0) to terminate the reaction. Immediately aft...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com