Primer pair, reagent kit and method for distinguishing brachymystax lenok from Hucho taimen
A technique of primer pair and leptospirosis, which is applied in biochemical equipment and methods, resistance to vector-borne diseases, measurement/inspection of microorganisms, etc., can solve problems such as difficult identification, loss of external morphological characteristics, and urgent identification
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0020] Embodiment 1, method establishment
[0021] 1. Sample genomic DNA extraction
[0022] Collect 5-10 scales of the sample fish, add 40 μl of lysate (Protease K 0.5mg / ml, Tris (pH8.0) 10mM, KCl 50mM, Tween 20 0.3% (w / v), NP-40 0.3% (w / v)), 55°C for 2h, 98°C for 10min to lyse, mix evenly after lysis, centrifuge at 1000-2000rpm for 1-2min, extract the supernatant, which is the crude genomic DNA extract.
[0023] 2. Primer design
[0024] One pair of specific primers were designed according to the ITS gene and the mitochondrial genome of the salmon and jerky salmon, and a total of 4 primers were designed. The primer sequences are as follows:
[0025] ITS-F: 5'-GTGCCTAACGGGGAAAGG-3' (SEQ ID NO.1)
[0026] ITS-R: 5'-AACATACAGCAAATGCCATCC-3' (SEQ ID NO.2)
[0027] mtDNA-F: 5'-TAAGCAAAATGGGCAAAACC-3' (SEQ ID NO.3)
[0028] mtDNA-R: 5'-GTTATTCCAAGCGCACCTTC-3' (SEQ ID NO.4)
[0029] The present invention compares the ITS gene of the salmon ITS on NCBI with the transcriptome ...
Embodiment 2
[0040] Embodiment 2, sample detection
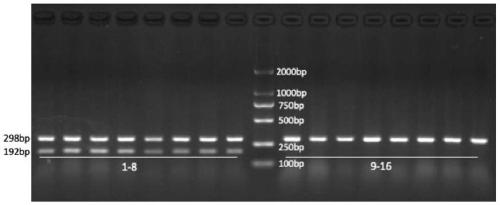
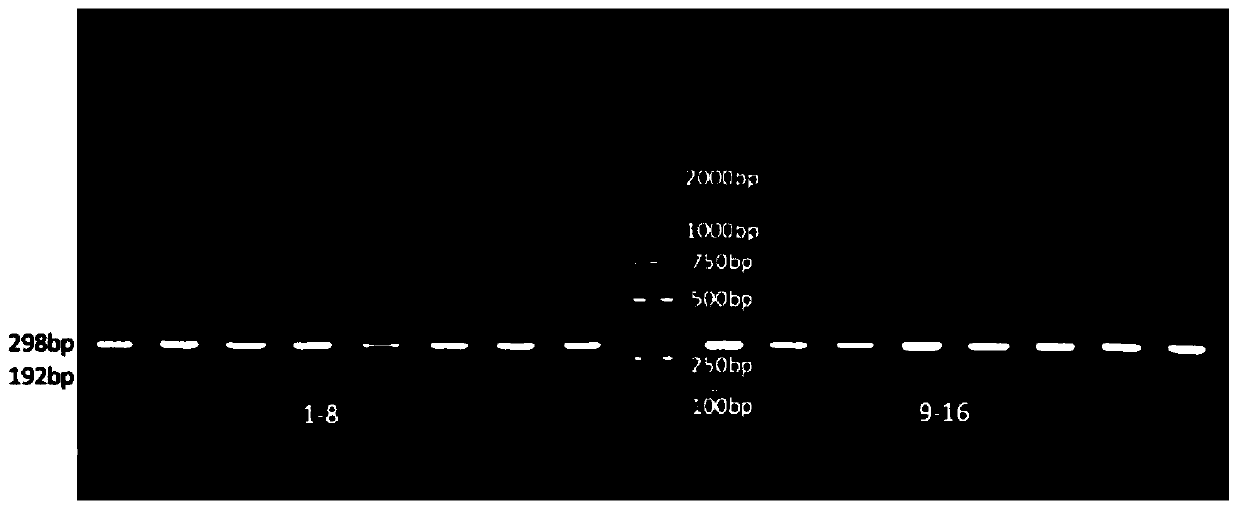
[0041] Using the identification method of Example 1, collect 50 adult fishes identified as wild leprosy salmon and salmon salmon in different watersheds according to their morphology, and get 5-10 scales of each adult fish for identification. According to the comparison between the electrophoresis results and the provided standard spectra of salmon salmon and salmon salmon, a specific band with a size of 298bp appeared in all 50 samples identified as salmon salmon by morphology, indicating that the samples were salmon salmon samples; 50 Two specific bands with a size of 298bp and a size of 192bp appeared in one sample of jerky salmon according to morphological identification, indicating that the sample was jerky salmon. The method is very sensitive, and fish eggs, juvenile fish and adult fish can all be distinguished, and the accuracy rate can reach 100%.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com