A Molecular Marker Related to Sheep Feed Conversion Rate and Its Application
A feed conversion rate and molecular marker technology, applied in the field of molecular marker preparation, can solve the problems of unclear genetic mechanism of sheep feed conversion rate and candidate genes that have not been reported yet, and achieve great practical application value, low-cost operation method, and cost reduction Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036] Example 1 Amplification of the PPARGC1B gene
[0037] (1) Primer design
[0038] Using sheep PPARGC1B gene DNA (GenBank accession number: NC_019462.2) as a template, a pair of primers M-F and M-R were designed using Primer5.0 software. The primer sequences are as follows
[0039] PPARGC1B:
[0040] M-F: 5'-ACCCATCAACGCTGCGAAAG-3' (SEQ ID NO: 2),
[0041] M-R: 5'-GCCAATGAGCCACCCTACAC-3' (SEQ ID NO: 3).
[0042] (2) Amplification and sequencing of PPARGC1B gene
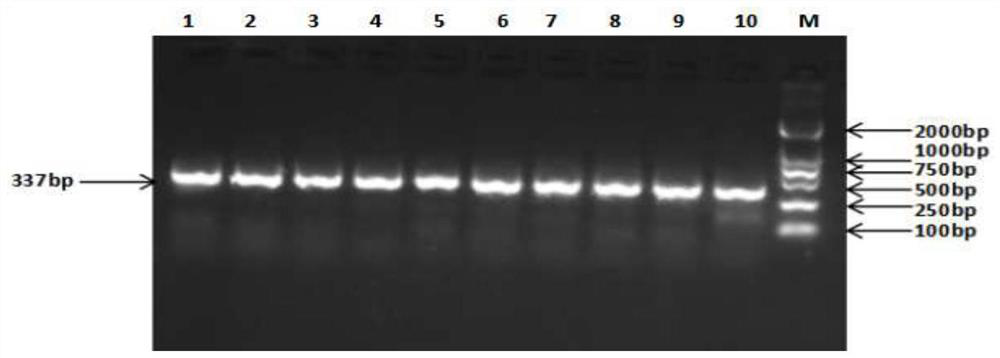
[0043] The total volume of the PCR reaction is 25 μL, including 1 μL of DNA template, 12.5 μL of 2×PCR Master Mix, 0.8 μL of upstream primer, 0.8 μL of downstream primer, ddH 2 O 9.9 μL. The PCR amplification program was: pre-denaturation at 94°C for 3 min, denaturation at 94°C for 30 s, annealing at 54.5°C for 30 s, extension at 72°C for 30 s, 35 cycles, and finally extension at 72°C for 10 min. The PCR reaction product was detected by 1.5% agarose gel electrophoresis, and the results showed that a 337bp s...
Embodiment 2
[0046] The establishment of embodiment 2 genotyping detection method
[0047] (1) Primer sequence design
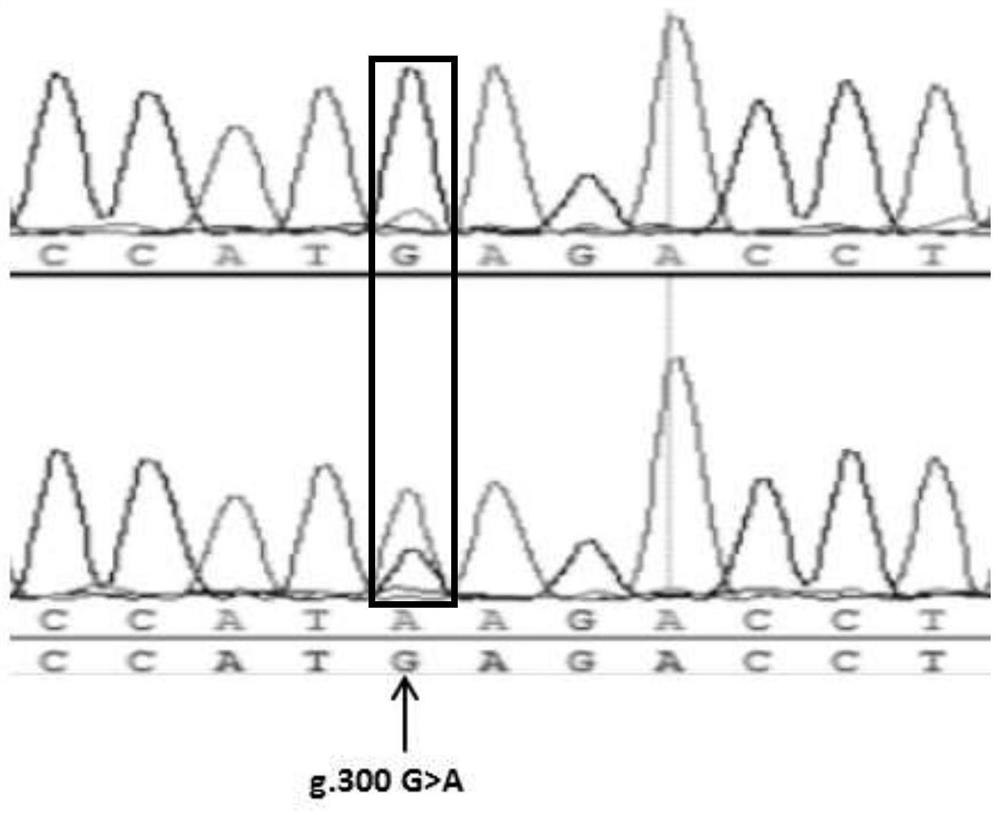
[0048] A pair of KASPar primers are designed for the G / A polymorphic site of the amplified fragment in Example 1, so as to be used for the specific detection of the polymorphic site, and the nucleotide sequence of the KASPar primer pair is:
[0049] Forward primer A1 for detection of AlleleA:
[0050] GAAGGTGACCAAGTTCATGCTTTCTAGAAATTGCTGACTTATTATTCCATA (SEQ ID NO: 4);
[0051] Forward primer A2 for detection of AlleleG:
[0052] GAAGGTCGGAGTCAACGGATTCTAGAAATTGCTGACTTATTATTCCATG (SEQ ID NO: 5);
[0053] Universal reverse primer C: CAATGAGCCACCCTACACCCAAC (SEQ ID NO: 6).
[0054] The above primers were synthesized by entrusting Beijing Sangong Bioengineering Co., Ltd., and the primers of each group in the KASPar primer pair were diluted to 10 μmol / L, and mixed according to the volume ratio of 12:12:30 (primer A1: primer A2: primer C) Evenly reserve.
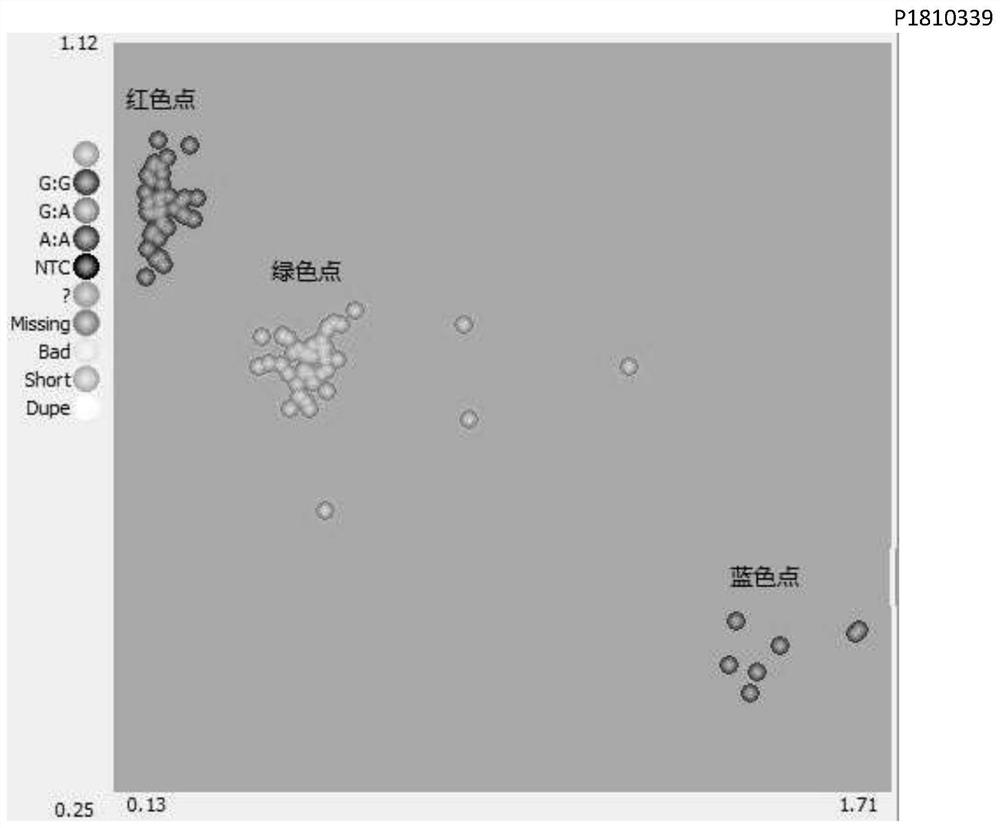
[0055] (2) DNA q...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com