Method for simultaneously detecting various intestinal tract viruses
A technology for enteroviruses and detection probes, which is applied in biochemical equipment and methods, microbial determination/inspection, DNA/RNA fragments, etc. Achieve strong specificity, avoid skewness, and high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
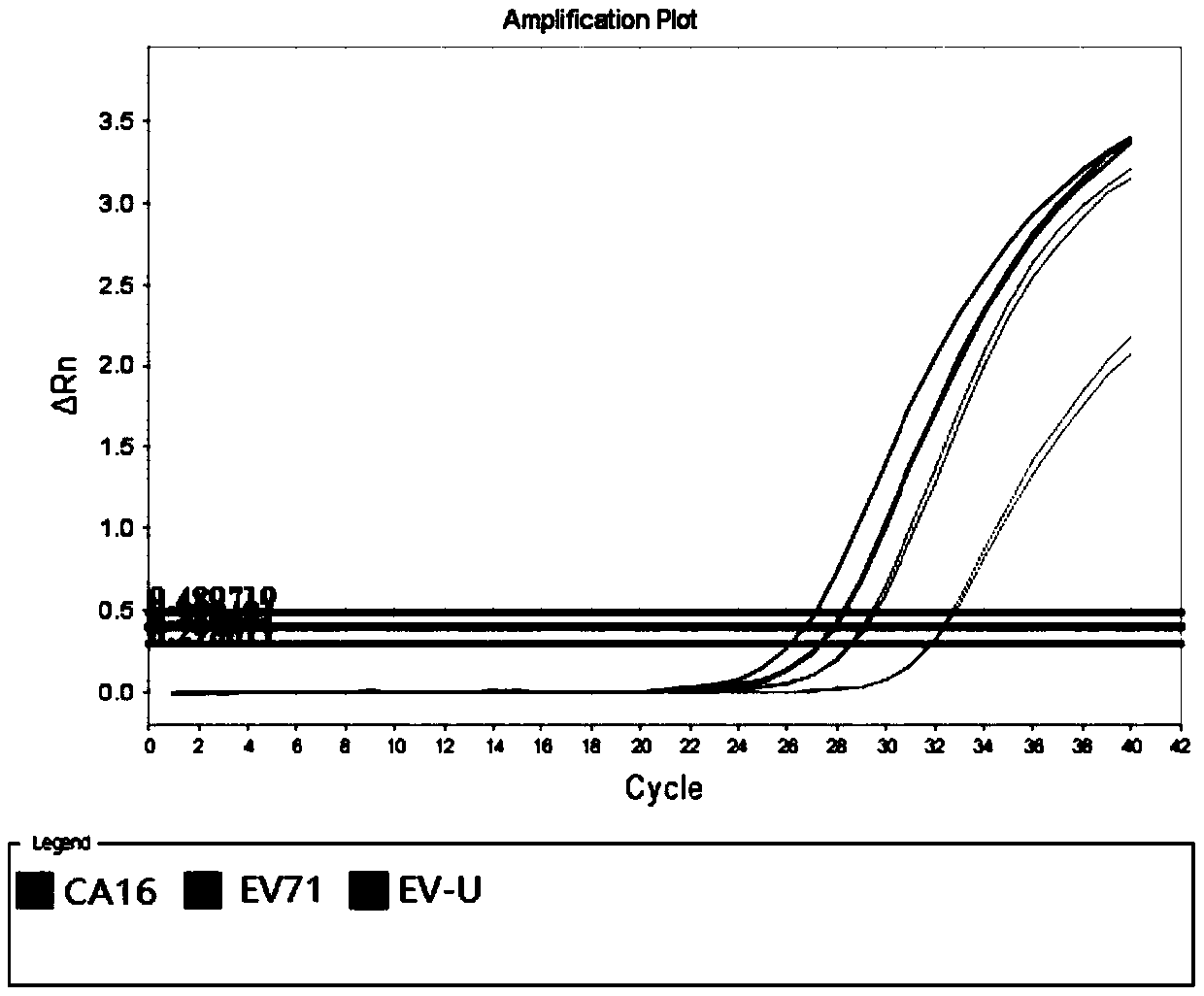
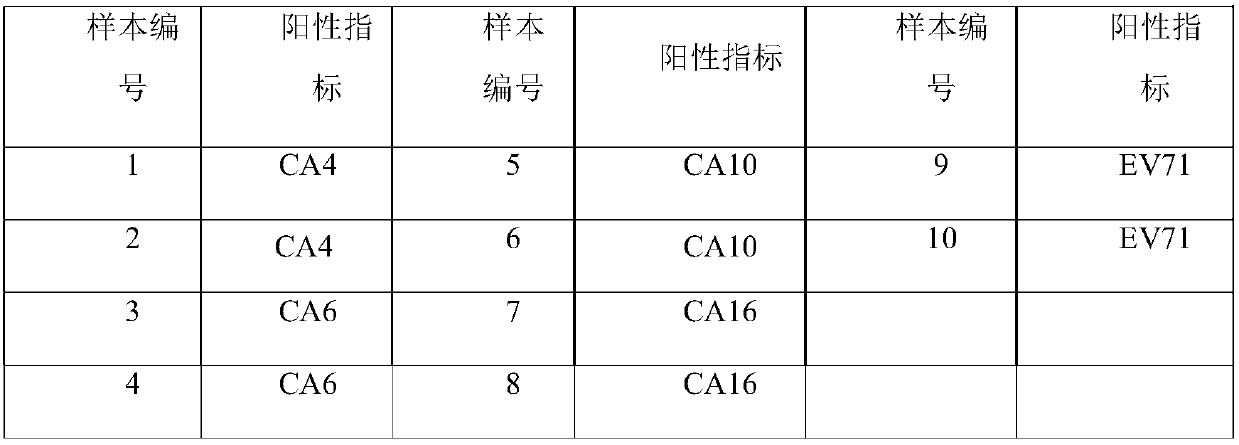
[0072] With the No. 001 / 002 / 003 sample described in Table 3 as a template, a negative control (using DEPC water as a template) was added at the same time, and the specific primers and universal primers for all pathogens described in Table 4 were used as amplification primers. The amplification method provided by the invention carries out multiple amplification, and the probes of EV-U, EV71, and CA16 in Table 4 are used as detection probes (wherein the EV-U detection probe 5' carries out FAM fluorescent labeling, and the 3' end carries out BHQ -1 fluorescent labeling; EV71 detection probe 5' is HEX fluorescent labeling, 3' end is BHQ-1 fluorescent labeling; CA16 detection probe 5' is Cy5 fluorescent labeling, 3' end is BHQ-2 fluorescent labeling), in multiplex Fluorescence quantitative PCR technology platform is the detection platform for detection.
[0073] The present embodiment is carried out according to the following steps:
[0074] 1. Enrichment and amplification
[007...
Embodiment 2
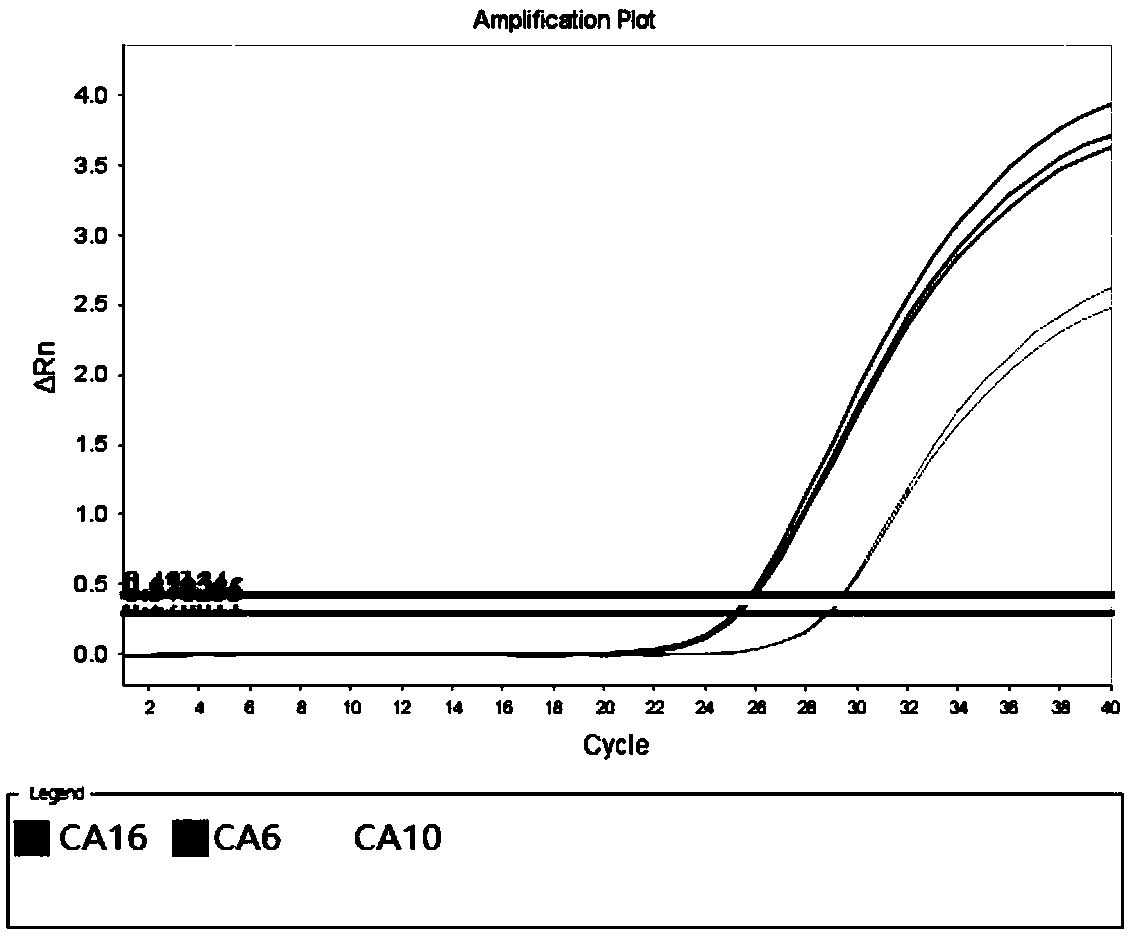
[0096] With No. 002 / 005 / 006 sample RNA described in Table 3 as a template, a negative control (using DEPC water as a template) was added simultaneously, and the specific primers and universal primers of all pathogens described in Table 4 were used as amplification primers for multiple amplification. In addition, the probes of CA6, CA10, and CA16 in Table 4 are used as detection probes (the 5' of the CA6 detection probe is fluorescently labeled with FAM, and the 3' end is fluorescently labeled with BHQ-1; the 5' of the detection probe of CA10 is fluorescently labeled with HEX labeling, BHQ-1 fluorescent labeling at the 3' end; Cy5 fluorescent labeling at the 5' end of the CA16 detection probe, and BHQ-2 fluorescent labeling at the 3' end), the multiplex fluorescent quantitative PCR technology platform was used as the detection platform for detection.
[0097] The present embodiment is carried out according to the following steps:
[0098] 1. Enrichment and amplification
[009...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com