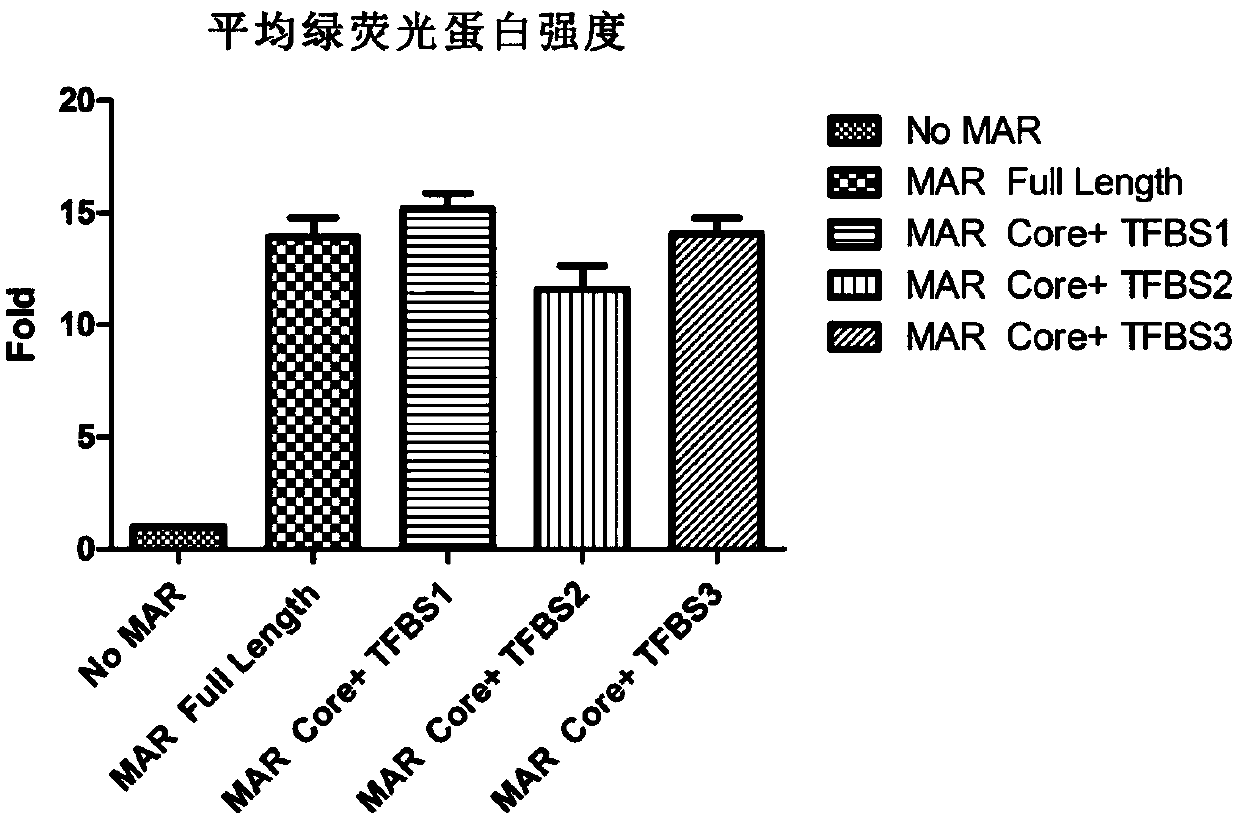
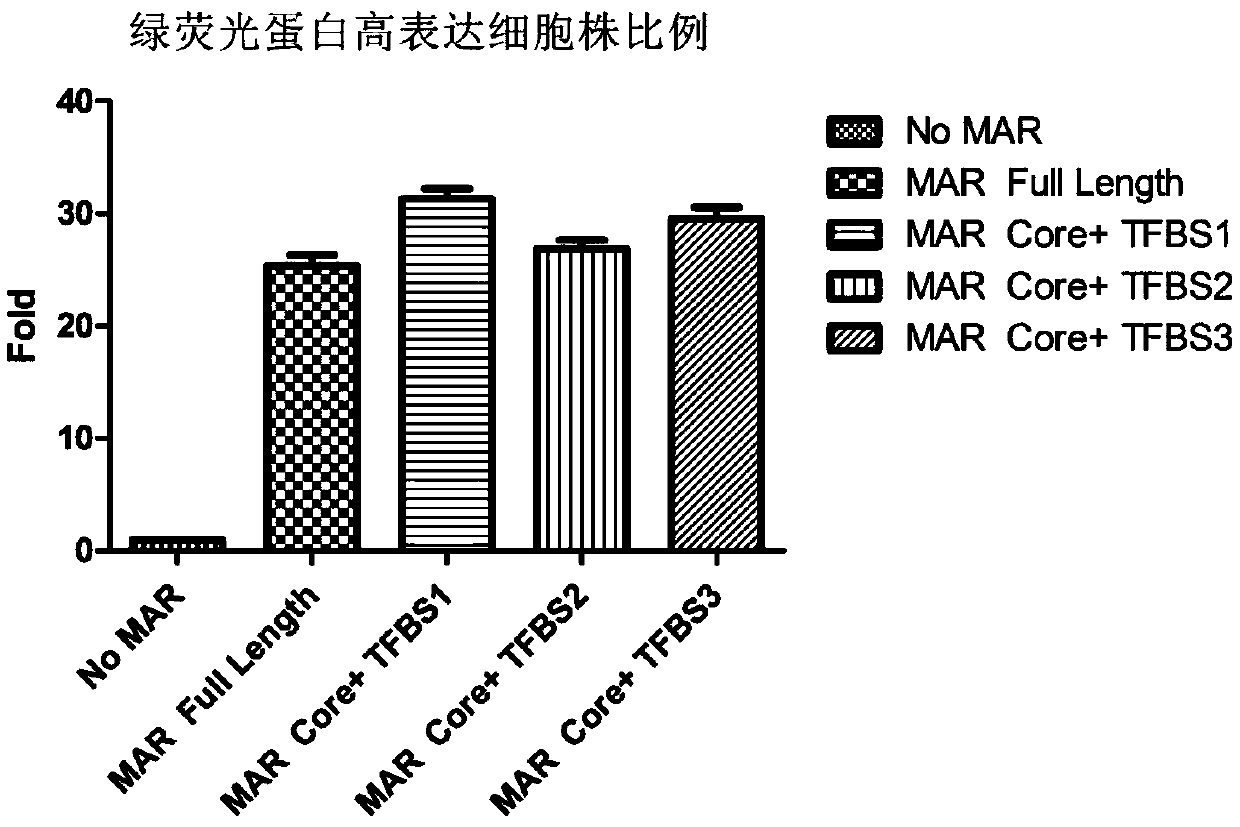
Novel animal cell expression vector containing MAR (Matrix Attachment Region) core fragment
A core fragment and expression vector technology, which can be applied to DNA/RNA fragments, cells modified by introducing foreign genetic material, and using vectors to introduce foreign genetic material. To achieve good stability and improve the effect of expression
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] Example 1 Introduction of Afe I and EcoR V restriction sites in pBudCE4.1
[0032] The Afe I and EcoR V restriction sites were introduced into the SV40 polyadenylation site and downstream of the BGH polyadenylation site by point mutation method. Use Quickchange Point Mutation Kit (purchased from Agilent Technologies) for point mutation. Introduce Afe I restriction site.
[0033] Forward primer: 5’-GGAAAACGATTCCGAAGC GCT AC-3’
[0034] Reverse primer: 5’-CCT TCT ATG AAA GGT AGC GCT TCG-3’
[0035] The PCR amplification conditions for site-directed mutagenesis are as follows: 1, 95°C for 30 seconds; 2, 95°C for 30 seconds; 3, 55°C for 1 minute; 4, 68°C for 5 minutes; 5, 2-4 repeat 18 cycles.
[0036] Add Dpn I enzyme to digest the template plasmid, and then transform XL1-Blue competent E. coli. The specific steps are as follows: 1. 100uL competent cells (purchased from Agilent Technologies), freshly prepared or stored at -20°C, are placed on ice, and the cells are gently suspend...
Embodiment 2
[0042] Example 2 Remove Zeocin resistance gene from pBudCE4.1 and add kanamycin resistance gene, PGK promoter, DHFR and SV40PA
[0043] The kanamycin resistance gene, PGK promoter, DHFR and SV40PA were PCR out of pCHO1.0 (purchased from LifeTechnologies), and loaded into pBudCE4.1 obtained in Example 1 through NheI and AfeI sites, and removed at the same time Zeocin resistance gene.
[0044] Forward primer: 5’-GTG AGCGCTTTAGAAAAACTCATCGAGCATC-3’
[0045] Reverse primer: 5'-GTG GCTAGCTAA GAT ACA TTG ATG AGT TTG-3'
[0046] Primestar HS high-fidelity DNA polymerase for PCR (purchased from TAKARA),
[0047] The specific steps are as follows: 1, 95°C for 3 minutes; 2, 98°C for 10 seconds; 3, 47°C for 15 seconds; 4, 72°C for 3 minutes; 5, 2-4 repeat 30 cycles; 6, 72°C for 5 minutes.
[0048] The modified new carrier is named PBCM1.0, and its structure diagram is attached Figure 5 Among them, the NheI, AfeI, FspI and EcoRV sites are the insertion sites of MAR core fragments with artificial t...
Embodiment 3
[0049] Example 3 Construction of an expression vector containing MAR core fragments with artificial transcription factor binding sites added at both ends of MAR core fragments with artificial transcription factor binding sites added at both ends (sequences such as SEQ ID NO: 1, SEQ ID NO: 2 Or SEQ IDNO: 3) synthesized by Shanghai Shenggong Biological Engineering Co., Ltd.
[0050] The MAR core fragment with artificial transcription factor binding sites at both ends can be inserted at the upstream FSPI site of PBCM1.0, or the downstream Afe I site of SV40PA, or the upstream Nhe I site of PEF-1a, or the downstream EcoRV site of BGH PA .
[0051] In the following example, the MAR core fragment with artificial transcription factor binding sites at both ends is inserted at the upstream FSPI site of PBCM 1.0 and the upstream Nhe I site of PEF-1a. The MAR core fragment sequence is as SEQ ID NO:1, SEQ ID NO: 2 or SEQ ID NO: 3.
[0052] The MAR core fragment with artificial transcription fa...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com