A probe and method for simultaneous detection of multiple microRNAs under constant temperature conditions based on DNA assembly
A probe and probe set technology, applied in DNA/RNA fragments, recombinant DNA technology, biochemical equipment and methods, etc., can solve problems such as complex system, and achieve the effect of simple amplification system and good accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0058] Example 1 A probe set for detecting microRNA
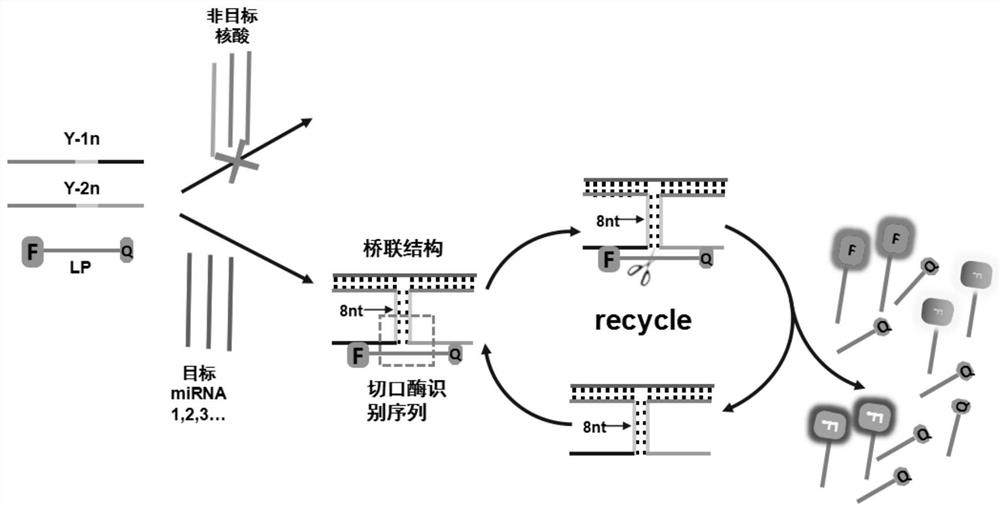
[0059] The probe set for detecting microRNA includes Y-1n, Y-2n and fluorescent probe LP; probes Y-1n, Y-2n, LP and target microRNA can self-assemble to form a stable "I"-shaped structure;
[0060] Part of the bases in the middle of the probe Y-1n are reverse-complementary paired with the middle part of the bases in Y-2n to form a stem sequence in the middle of the "I" shape; the length of the stem sequence is 7 to 9 bp; the The stem sequence contains the recognition site for the nicking endonuclease.
[0061] The other part of the base sequence of the probes Y-1n and Y-2n is reverse-complementary paired with the two ends of the target microRNA respectively, forming a "one"-shaped head at one end of the "I" shape;
[0062] There are also base sequences in the probes Y-1n and Y-2n that can undergo reverse complementary pairing with the sequences at both ends of the fluorescent probe LP, forming a "one"-shaped head at the ot...
Embodiment 2
[0067] Embodiment 2 A method for detecting microRNA
[0068] method:
[0069] A target miRNA (miR-373) was selected and a probe was designed by NUPACK software, and DNA probes Y-11, Y-21 and fluorescent probe LP partially complementary to the sequence were synthesized. The specific sequences of each probe are shown in Table 1 shown.
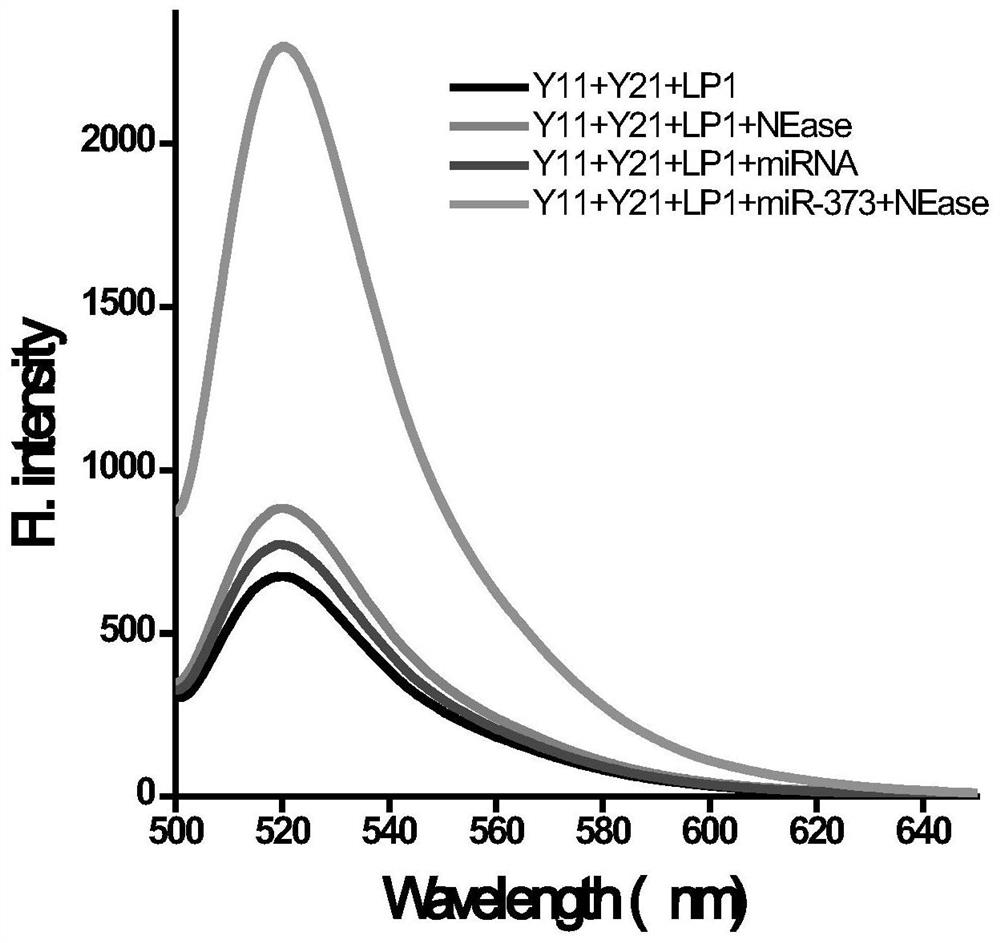
[0070] Mix Y-11, Y-21, LP, Nt.BstNBI, Nt.BstNBI buffer and target miRNA, and react at 45°C for 30 minutes; simultaneously detect the change of fluorescence intensity.
[0071] At the same time, three groups of control experiments were set up:
[0072] Y-11+Y-21+LP1 group: when only Y-11, Y-21 and LP exist in the reaction system of the control group, the nicking endonuclease Nt.BstNBI and target miRNA (miR-373) are not included, other The conditions were the same as the above experimental group.
[0073] Y-11+Y-21+LP1+NEase group: the reaction system of the control group does not contain the target miRNA (miR-373), and other conditions are the...
Embodiment 3
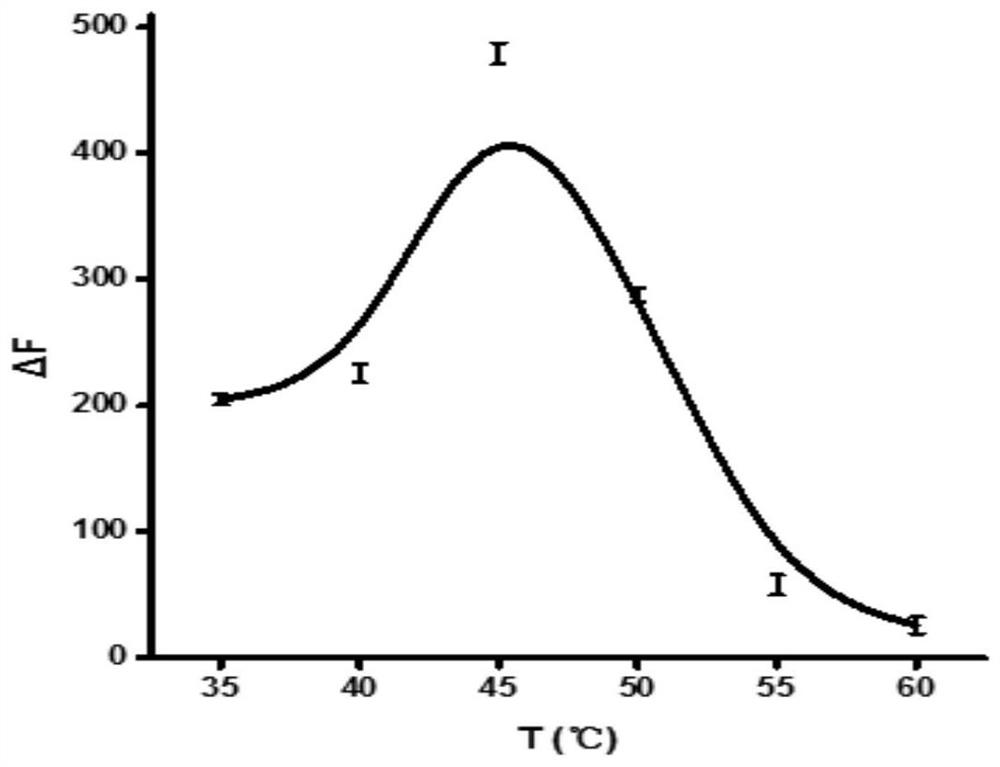
[0081] The investigation of embodiment 3 reaction temperature
[0082] method:
[0083] According to the physical and chemical properties of the restriction endonuclease Nt.BstNBI, its catalytic activity is higher at about 55°C, and its activity will be significantly reduced if the reaction temperature is too high or too low. In addition, the melting temperature of the "I"-shaped bridge structure in the system also has a great impact on the reaction. When the reaction temperature is too high, it will be difficult to form this structure, thus affecting the enzymatic digestion cycle amplification process. In this system, keeping other conditions constant, 35, 40, 45, 50, 55 and 60°C were selected as reaction temperatures to investigate the effect of temperature on the fluorescence intensity of the reaction system. The specific operation method is: mix Y-11, Y-21, LP, Nt.BstNBI, Nt.BstNBI buffer and target miRNA (miR-373), respectively, at 35, 40, 45, 50, 55 and 60 ° C The reac...
PUM
| Property | Measurement | Unit |
|---|---|---|
| recovery rate | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com