CRISPR-Cas based isothermal nucleic acid detection method and kit
A detection method and nucleic acid technology, applied in the field of isothermal nucleic acid detection and kits based on CRISPR-Cas, can solve the problems that RNA detection cannot realize one-step single-tube detection, the RPA method has complex components, and increases the difficulty of operation, etc., to achieve low false positives Positive, easy-to-step, high-sensitivity effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
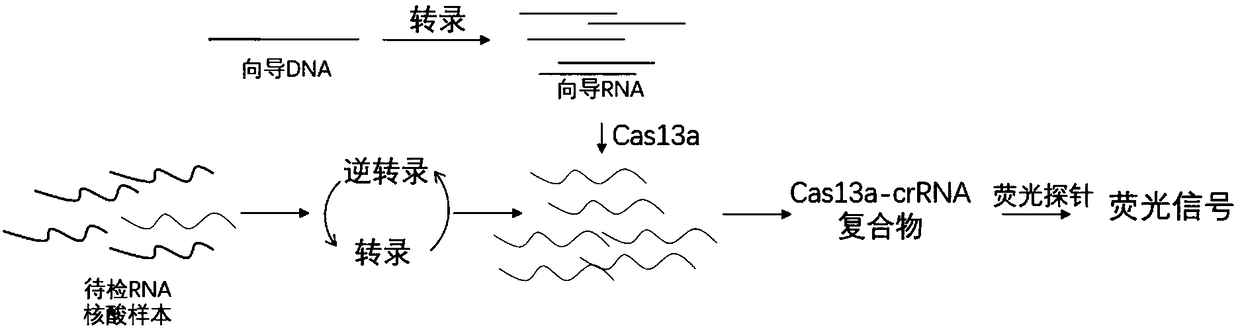
[0059] Example 1: Detection of RNA targets using the present invention
[0060] RNA (Target1) is selected as the target sequence, and the Target1 sequence is: aucgauagucuagucaucguacguagcuagucagucaauaucgauca;
[0061] The preparation method of the target RNA is to synthesize the forward primer (T7-F) taatacgactcactatag of the forward primer (T7-F) taatacgactcactatag of the reverse complementary long primer (Target1-primer-R) tgatcgatattgactgactagctacgtacgatgactagactatcgatccctatagtgagtcgtattaT7 comprising the T7 sequence, and make the DNA incomplete double strand by annealing of the double primers. T7 transcriptase was reacted overnight at 37 degrees, and then purified using the ZYMO RNA CLEAN&CONCENTRATION kit (Zymo Research Company) to obtain target RNA, which was stored at -20 degrees or -80 degrees after preparation.
[0062] Preparation of guide DNA: Synthesize the reverse complementary long primer crDNA-target1-R:agctacgtacgatgactagacGTTTTAGTCCCCTTCGTTTTTGGGGTAGTCTAAATCCCC...
Embodiment 2
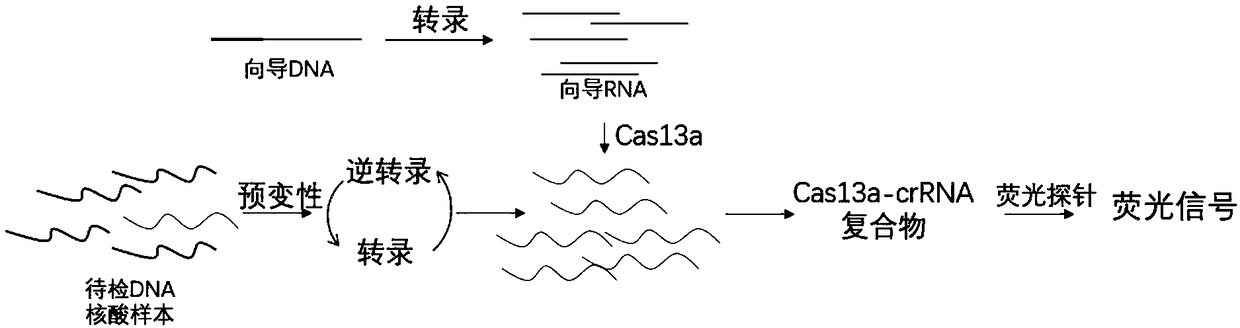
[0067] Embodiment 2: Using the present invention to detect single-stranded DNA targets
[0068] DNA (Target2) is selected as the target sequence, and the Target2 sequence is: atcgatagtctagtcatcgtacgtagctagtcagtcaatatcgatca.
[0069] The method for preparing the target single-stranded DNA is to synthesize a primer (Target2-primer-F): atcgatagtctagtcatcgtacgtagctagtcagtcaatatcgatcacccaatgtgacatcgctga, dissolve in water and dilute to 100uM.
[0070] Preparation of guide DNA: Synthesize the reverse complementary long primer crDNA-target2-R containing the T7 sequence: agctacgtacgatgactagacGTTTTAGTCCCCTTCGTTTTTTGGGGTAGTCTAAATCCCCTATAGTGAGTCGTATTAA T7 forward primer (T7-F) taatacgactcactatag, and make the DNA incompletely double-stranded by annealing the double primers. Store at -20°C or -80°C after preparation.
[0071] Amplification and detection reaction: Add the single-stranded DNA to be detected into the reaction system, the reaction system includes, buffer (trishydrochloride, 50...
Embodiment 3
[0074] Example 3: Detection of double-stranded DNA targets using the present invention
[0075] Select DNA (Target3) as the target sequence, and the Target3 sequence is:
[0076] atcgatagtctagtcatcgtacgtagctagtcagtcaatatcgatcagacatcgaggagatcaaaacccagaaggtccgcatcgaaggc.
[0077] The preparation method of the target DNA is to synthesize a reverse complementary long primer (Target3-primer-R)
[0078] gccttcgatgcggaccttctgggttttgatctcctcgatgtctgatcgatattgactgactagctacgtacgatgactagactatcgat, and forward primer (Target3-primer-F)
[0079] atcgatagtctagtcatcgtacgtagctagtcagtcaatatcgatcagacatcgaggagatcaaaacccagaaggtccgcatcgaaggc, the primers were mixed and annealed, and stored at -20 degrees or -80 degrees after preparation.
[0080] Preparation of guide DNA: Synthesize the reverse complementary long primer crDNA-target3-R containing the T7 sequence: agctacgtacgatgactagacGTTTTAGTCCCCTTCGTTTTTTGGGGTAGTCTAAATCCCCTATAGTGAGTCGTATTAA T7 forward primer (T7-F) taatacgactcactatag, and make ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com