Method for rapidly establishing animal model of Parkinson's disease through substantia nigra gene knockout
A technology for establishing methods and animal models, applied in the biological field, can solve problems such as increasing the difficulty of research, and achieve the effect of stable phenotype
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0059] The transformation of embodiment 1 expression vector
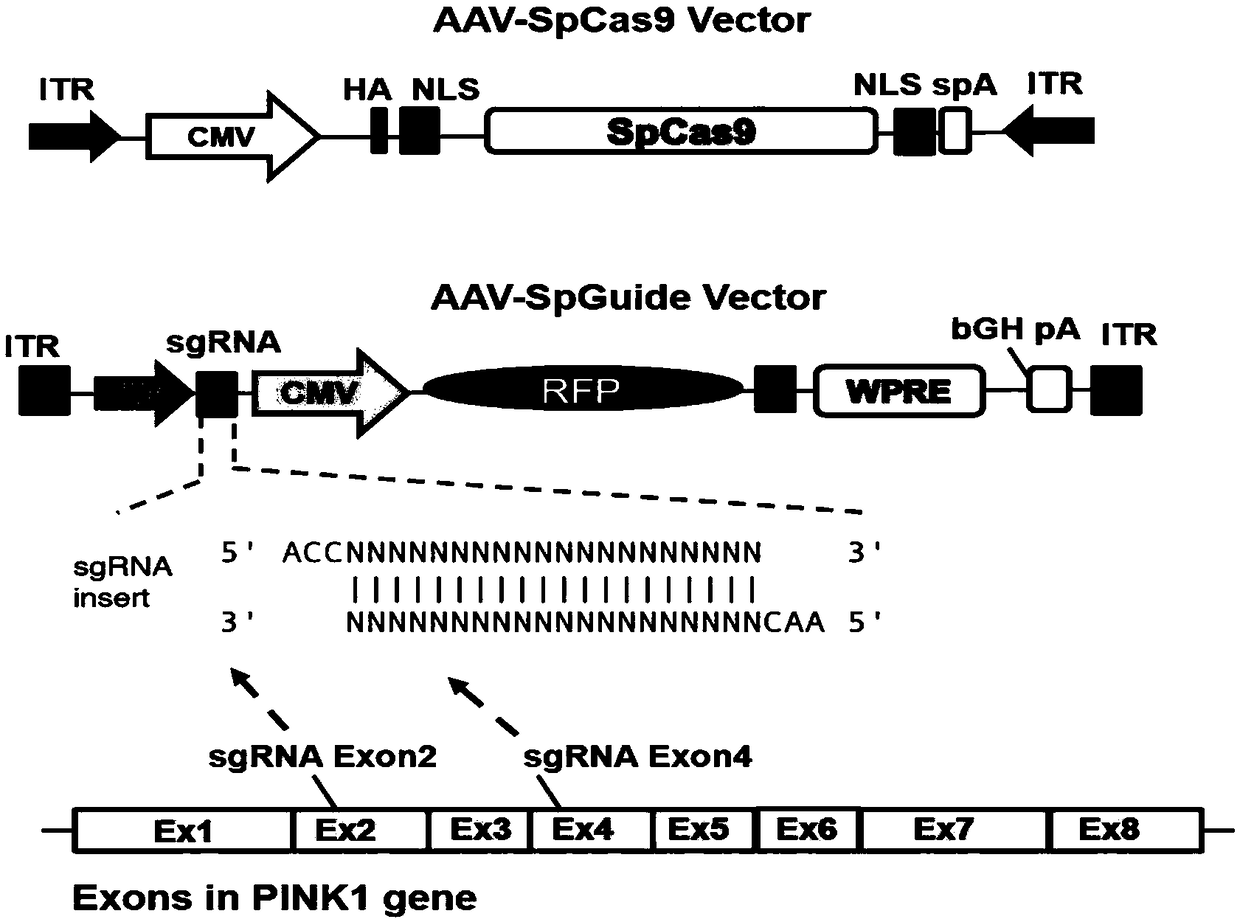
[0060] Transform viral vectors expressing target plasmids such as figure 1 , the specific construction process method is as follows (relevant specific operations can be realized by conventional technical means in the field):
[0061] 1. Use the Xba1 and AgeI restriction sites on the pAAV-pMecp2-SpCas9-spA (purchased from Addgene, px551, plasmamid#60957) backbone plasmid to delete the Mecp2 promoter sequence, and connect the Xba1 and AgeI restriction sequences The CMV promoter sequence, so as to realize the expression of SpCas9 in each cell in the brain, and obtain AAV-Cas9.
[0062] 2. For the transformation of the AAV-U6sgRNA-hSyn-GFP (Addgene, px552, plasmamid#60958) plasmid, the hSyn sequence in the original vector was transformed into the CMV promoter sequence through ApaI and BamHI, and cut by BamHI and EcoRI Point to replace the GFP sequence with the RFP sequence to get the AAV-U6sgRNA-CMV-RFP vector.
[00...
Embodiment 2
[0064] Example 2 Constructing the sgRNA vector of CRISPR / Cas9 containing monkey PINK1 gene target sequence
[0065] 1. Design of sgRNA sequence
[0066] In order to screen the PINK1 sgRNA sequence capable of gene editing and with small off-target effects, the applicant used NCBI's Blast software to screen the monkey PINK1 protein coding region gene (SEQ ID NO.1) with low off-target effects and consistent with the CRISPR / Cas9 binding genome Featured (5'-G(19N)-NGG3') or (5'-CCN(19N)-C-3') target sequence sites.
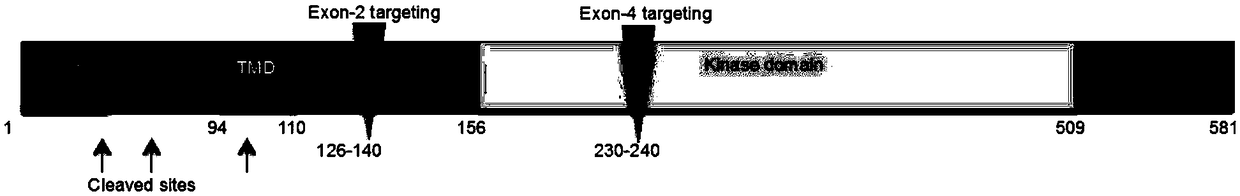
[0067] Inventors in monkey PINK1 gene No. 2 and exon No. 4 ( figure 2 ) respectively designed multiple sgRNAs, namely: PINK1exon2-T1, T2, T3 ( image 3 ) and PINK1exon4-T1, T2, T3 ( Figure 4 ), wherein the PINK1 exon2-T target sequence contains a specific Hpy188I restriction site sequence to facilitate the detection of mutations. The specific sequence information of sgRNA is shown in Table 1:
[0068] Table 1 sgRNA sequence information
[0069]
[0070]
...
Embodiment 3
[0111] Example 3 Virus Packaging and Construction of Animal Models
[0112] The viral vectors AAV-sgRNA-PINK1 Exon2 and AAV-sgRNA-PINK1 Exon4 transformed and constructed in Example 2 and the AAV-Cas9 constructed in Example 1 were used for virus packaging. For specific steps, see the literature Yang et.al., J Clin Invest, 2017 (doi:10.1172 / JCI92087), and injected into the substantia nigra of monkey brain through stereotaxic injection method, the operation method is as follows Figure 9 shown. A total of 11 cynomolgus monkeys were used to construct the animal model, of which 6 were male and 5 were female; 3 were 3-year-old animals, 4 were 8-year-old animals; and 4 were 12-year-old animals.
[0113] The specific injection scheme is as follows: stereotaxic positioning of the substantia nigra of the monkey brain through nuclear magnetic resonance scanning images (MRI), and the parameters are based on the actual measurement of the MRI. Subsequently, 12 μL was injected into the right...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com