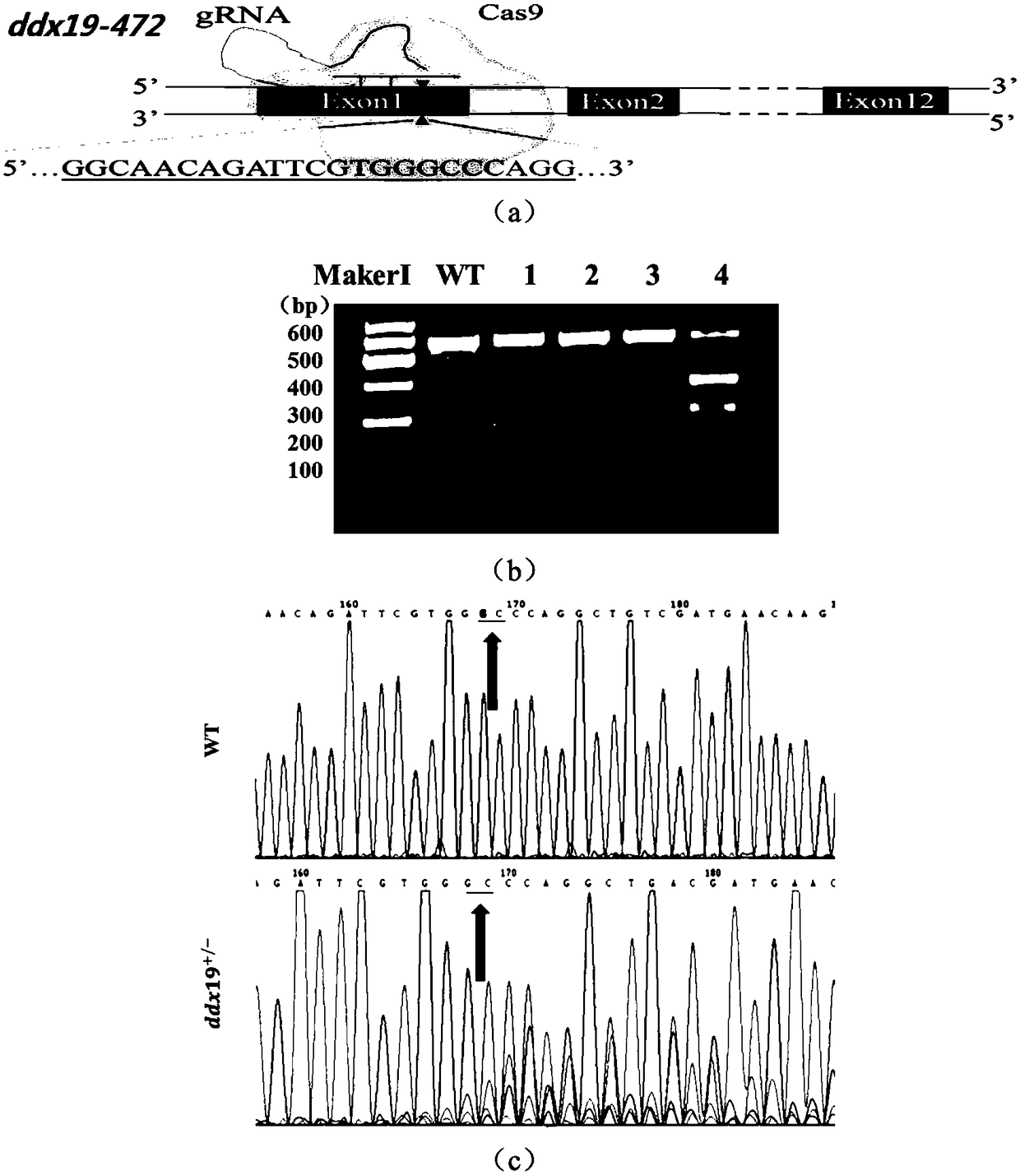
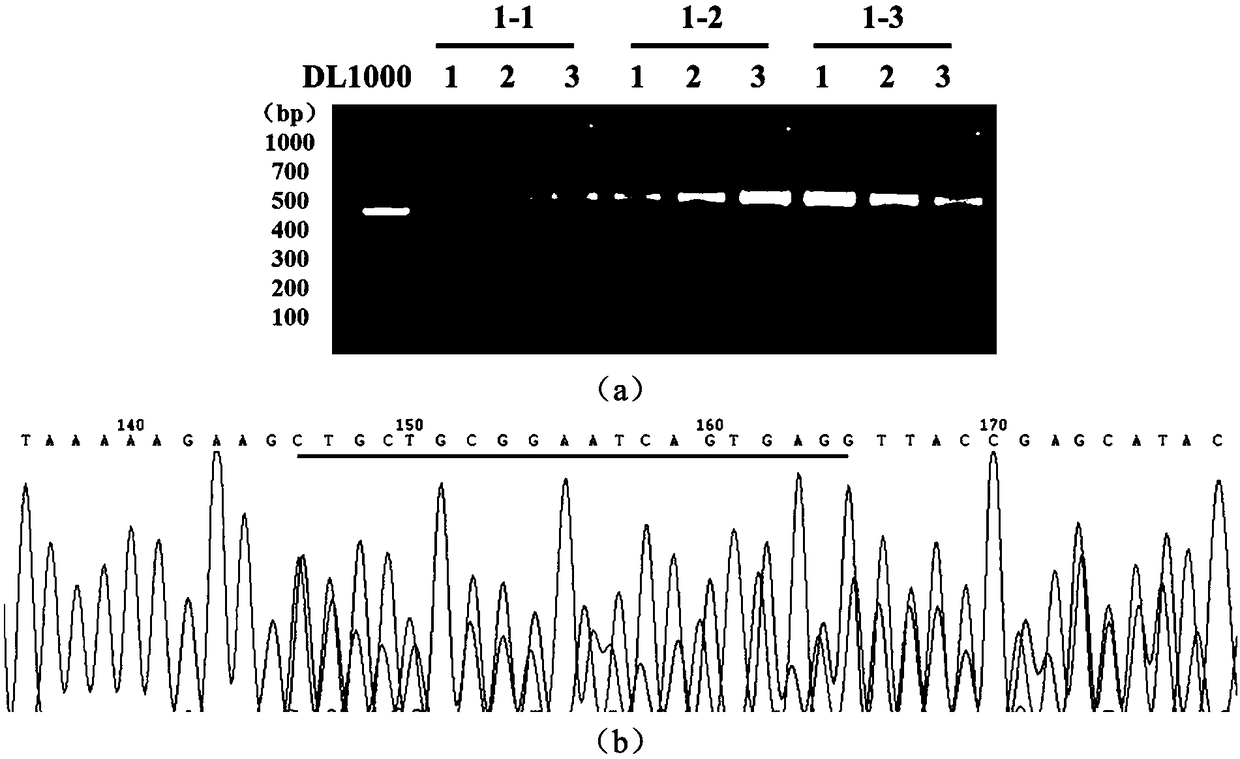
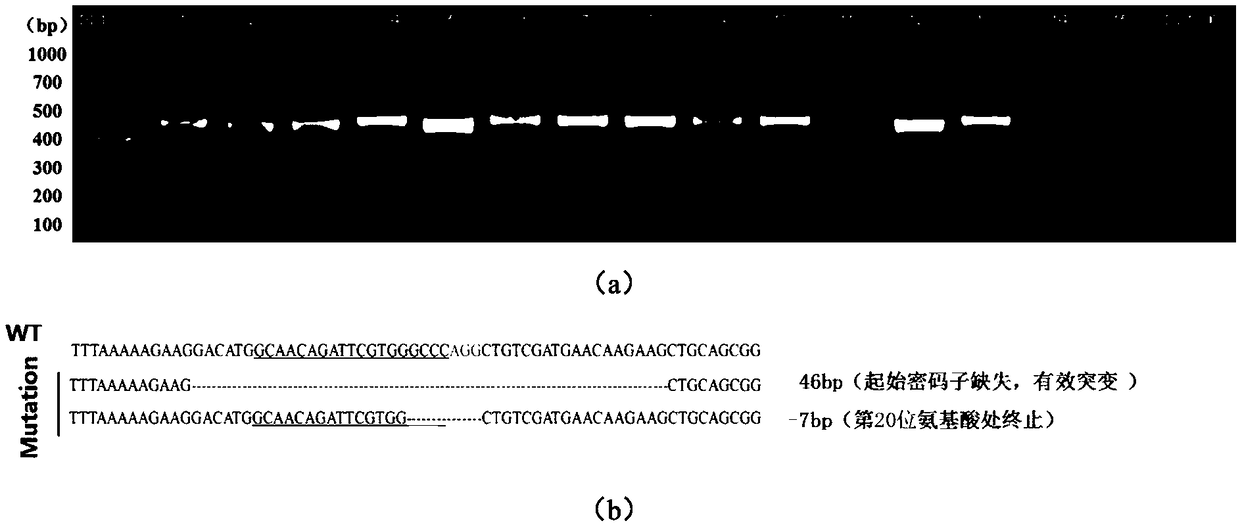
Preparation method of ddx19 gene-deleted zebrafish mutant
A gene deletion, t7-ddx19-sfd technology, applied in the field of molecular biology, can solve the problems of different types of mutants, different sizes of encoded proteins, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
[0058] 1 Materials and equipment
[0059] 1.1 Experimental fish
[0060] The zebrafish used in this experiment were all AB strains, which were purchased from the Zebrafish Platform of the Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences.
[0061] 1.2 Plasmid
[0062] The pUC19-gRNA scaffold plasmid is derived from literature: Chang N, Sun C, Gao L, Zhu D, Xu X, ZhuX, Xiong JW, Xi JJ. Genome editing with RNA-guided Cas9nuclease in zebrafishembrvos, Cell Res, 2013, 23(4 ): 465-472.
[0063] The pUC19-gRNA scaffold plasmid template sequence used in gRNA product synthesis is:
[0064] GTTTTAGAGCTAGAAATAGCAAGTTAAAATAAGGCTAGTCCGTTATCAACTTGAAAAAGTGGCACCGAGTCGGTGCTTTTTTT (SEQ ID NO. 7).
[0065] 1.3 Main reagents
[0066] DNA Clean&Contentrator-5 (ZYMO RESEARCH, D4004), common DNA purification kit (TIANGEN, DP204-03), T7in vitro Transcription Kit (Ambion, AM1314), ethanol (absolute ethanol) (Sinopharm Chemical...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com