Detection module and method
A detection module, nucleic acid sequence technology, applied in the direction of biochemical equipment and methods, microbial determination/inspection, etc., can solve problems such as inability to perform on-site detection, time-consuming, and little universal detection.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0093] Example 1, Nucleotide sequences used in EXPAR tandem digestion HCR detection module and design method thereof
[0094] (1) Nucleotide sequence used by EXPAR
[0095] The nucleotide sequences used in the EXPAR designed in this embodiment are shown in Table 1.
[0096] Table 1
[0097]
[0098] (2) Nucleotide sequences of traditional HCR and digested HCR
[0099] The nucleotide sequences used in traditional HCR and digested HCR designed in this example are shown in Table 2.
[0100] Table 2
[0101]
[0102] In Table 2, hairpin H1 is hairpin 1, hairpin H2 is hairpin 2, which is used for traditional HCR; D-H1 is digestion hairpin 1, and D-H2 is digestion hairpin 2, which is used for digestion type HCR.
[0103] (3) Design method of EXPAR serial digestion type HCR general module
[0104] In this embodiment, sequence X is used to represent the target sequence to be detected. In the actual detection process, the primer sequence can be designed according to the tar...
Embodiment 2
[0127] Embodiment 2, establishment and optimization of EXPAR reaction system, reaction conditions
[0128] (1) Establishment of EXPAR reaction system and reaction conditions
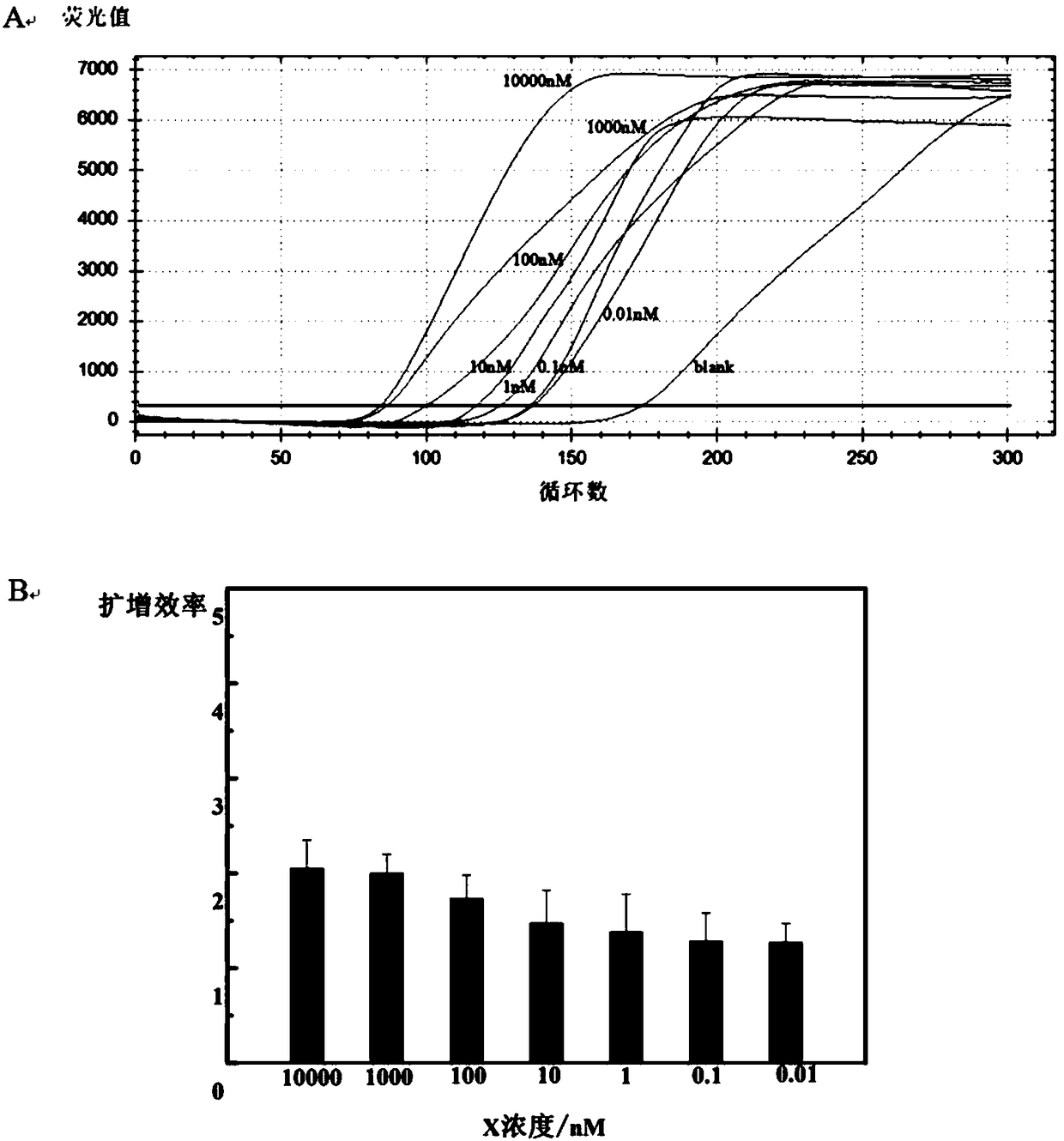
[0129] The EXPAR reaction system established in this example is divided into two systems, A and B, as shown in Table 4 and Table 5 respectively. The established EXPAR reaction system was verified by real-time PCR method.
[0130] System A is shown in Table 4.
[0131] Table 4
[0132]
[0133]
[0134] System B is shown in Table 5.
[0135] table 5
[0136]
[0137] EXPAR reaction conditions:
[0138] Add the samples except SYBR Green I in system A, mix well, put in a reaction tube, and heat at 55°C for 5min. After taking it out, add SYBR Green I (avoid light as much as possible). Add system B to system A, centrifuge to mix the reactants, and place them in a real-time quantitative PCR instrument for reaction. The design program is 55°C, 250 cycles.
[0139] Experimental results such as f...
Embodiment 3
[0148] Embodiment 3, establishment and optimization of digested HCR
[0149] (1) Self-assembly effect of digested HCR, verification of priming efficiency and establishment of self-assembly reaction conditions
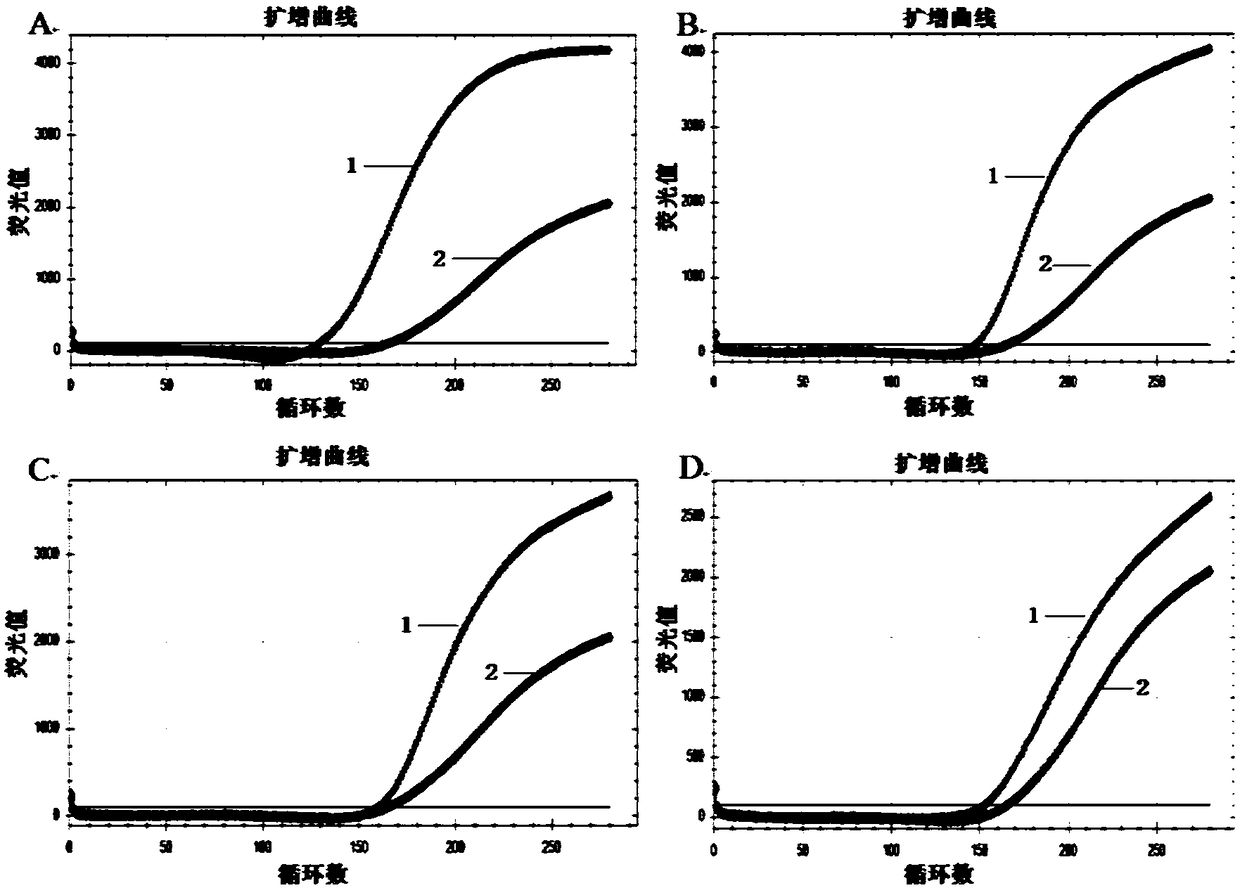
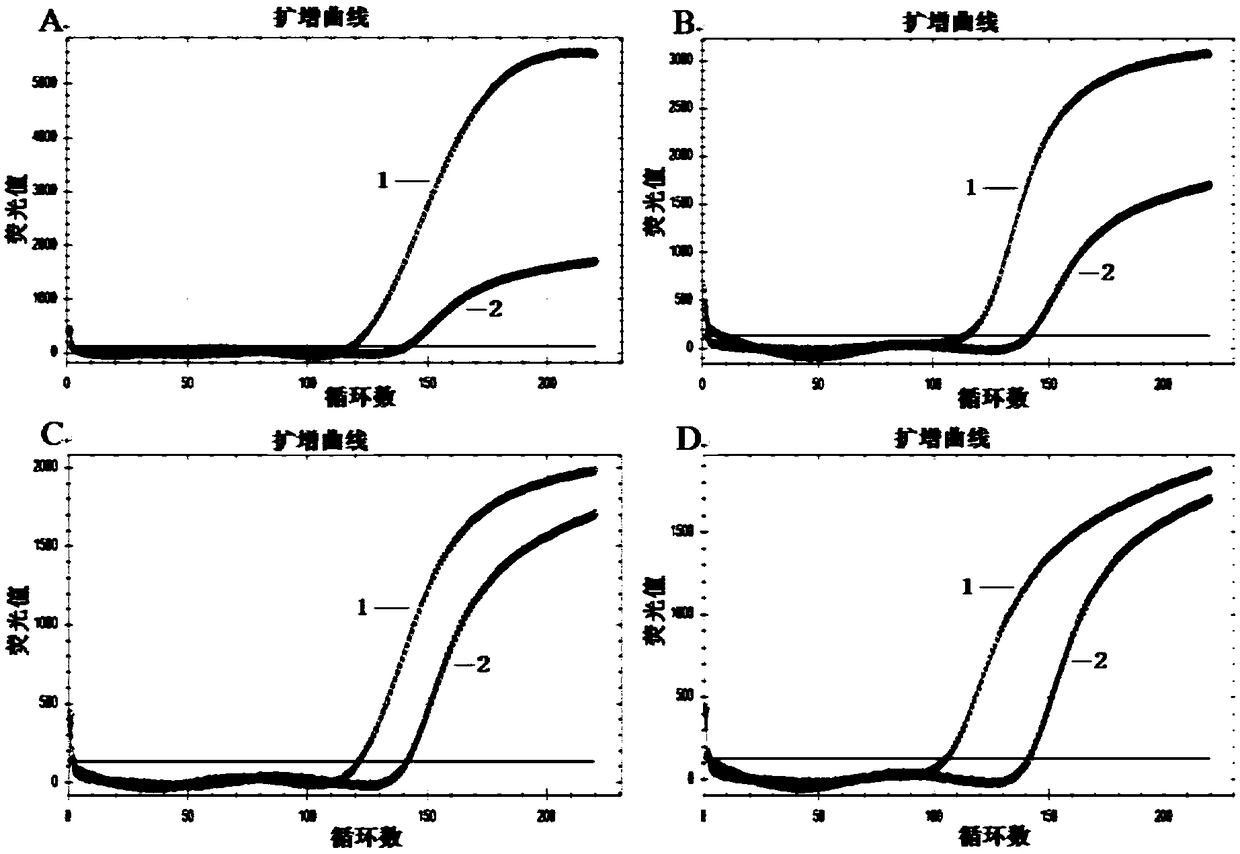
[0150] The hairpin H1 and hairpin H2 used for traditional HCR in Table 2 of Example 1 were used as the control group (hairpin H1 is abbreviated as H1 in the following description, and hairpin H2 is abbreviated as H2), and D-H1 and D- H2 is used as the experimental group, when the concentration of each hairpin structure is 100nM, the promoter concentration in Table 2 of Example 1 is 10nM, and the self-assembly reaction time is 30min, 1h, 90min, 2h, 150min, 3h, 4h respectively , the self-assembly effect and priming efficiency of the experimental group and the control group.
[0151] 1) Self-assembly reaction system: each of the two hairpin structures is 100nM, the promoter is 10nM, and the reaction buffer (8.0mMNa 2 HPO 4 ,2.5mM NaH 2 PO 4 ,2.0mM MgCl 2 , and 0.15mM...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com