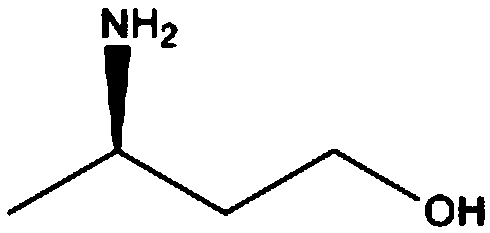
Transaminase derived from actinomyces, mutant, recombinant bacteria and application
A transaminase and mutant technology, which is applied to the transaminase, mutant, recombinant bacteria and application fields derived from actinomycetes, can solve the problems of poor selectivity, unsuitable for large-scale industrial production of column chromatography method, high price, etc. The effect of improved conversion rate, excellent catalytic activity, and low industrial cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0028] Example 1 Construction of transaminase-containing genetically engineered bacteria
[0029] The transaminase gene (nucleotide sequence shown in SEQ ID NO.1, amino acid sequence shown in SEQ ID NO.2) derived from actinomycete (Actinobacteriasp) was selected for whole gene synthesis, connected with plasmid pET-28b, and transferred into Escherichia coli DH5α competent cells were spread on LB plates containing kanamycin (50 μg / ml), cultured overnight at 37°C, and positive transformants were picked and identified and sequenced. Inoculate the verified positive single clone into 5 mL of LB liquid medium containing 50 μg / mL kanamycin, culture overnight at 37°C, extract the plasmid, and after re-verification, transfer the recombinant expression vector into Escherichia coli BL21(DE3) Among the strains, the recombinant strain E.coli BL21(DE3) / pET28b-TA0 (ie, the parent strain) was obtained, and cultured overnight at 37° C. on an LB plate containing kanamycin (50 μg / ml). Pick a sin...
Embodiment 2
[0030] Example 2 Obtaining of transaminase mutants and construction of genetically engineered bacteria containing mutants
[0031] 1. Construction of mutant library
[0032] Using the recombinant expression vector pET28b-TA0 obtained in Example 1 as a template, the mutant sequence was obtained by error-prone PCR amplification. Amplification primers are (5'GCTGA GGATCC ATGACCATCTCTAAAGACAT3') and (5'GCATC AAGCTT TCAGTATTCGATAGCTTC3').
[0033] Amplification system: 50μl Reaction system: 10xTaq polymerase buffer: 5μl; Mg 2+ (25mM): 2-8μl; Mm 2+ (25mM): 2-8μl; 10mM dNTP mixture (dATP, dCTP, dGTP and dTTP each 2.5mM) 4μL; concentration of 50μM upstream primer, downstream primer 1μL each, DNA template: 1μL; Taq DNA polymerase: 10U; Top up the system with steamed water.
[0034] The PCR reaction conditions were as follows: pre-denaturation at 95°C for 1min, followed by a temperature cycle of 95°C for 10s, 56°C for 90s, and 72°C for 1min, a total of 30 cycles, and finally an ...
Embodiment 3
[0038] Embodiment 3 Preparation of transaminase wild-type and mutant catalysts
[0039] 1) Slant culture: Inoculate recombinant bacteria E.coliBL21(DE3) / pET28b-TA0 and recombinant bacteria E.coliBL21(DE3) / pET28b-TA1 containing transaminase wild-type gene and transaminase mutant gene into card containing 50 μg / ml respectively. The LB medium of namycin was cultured at 37°C for 16h to obtain slant bacteria; the mass final concentration of the LB medium consisted of: peptone 10g / L, yeast extract 5g / L, sodium chloride 10g / L, 1.5 % agar, the solvent is deionized water, pH 7.0, and 50 μg / ml kanamycin is added before use.
[0040] 2) Seed culture: inoculate the slant bacteria into LB liquid medium containing 50 μg / ml kanamycin, cultivate at 37°C for 8-10 hours, and obtain seed liquid; the final concentration of the LB liquid medium is composed of: peptone 10g / L, yeast extract 5g / L, sodium chloride 10g / L, solvent is deionized water, pH 7.0, add 50μg / ml kanamycin before use.
[0041]...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com