LAMP (loop-mediated isothermal amplification) detection primer of fusarium proliferatum and application of LAMP detection primer
A technology for detection of Fusarium exfoliates and primers, which is applied in the fields of identification and control, and detection of crop diseases, can solve the problems of research reports on the rapid detection of Fusarium exfoliates, and achieve accurate and reliable detection results, strong specificity, and detection short cycle effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0053] The design of the specific primer composition for the LAMP detection of Fusarium exfoliates: using Clustal X software, through the Fusarium exfoliates and other Fusarium ( Fusarium spp.) EF-1alpha Gene sequences were compared and analyzed, and a set of nucleotide sequences shown in SEQ ID NO.1 to SEQ ID NO.6 was designed using the online LAMP primer design software Primer Explorer V5 software (http: / / primerexplorer.jp / lampv5e) The layered Fusarium-specific LAMP primer composition is specifically composed of a pair of outer primers F3 / B3, a pair of inner primers FIP / BIP and a pair of loop primers Loop F / Loop B.
[0054] The specific primer sequences are shown below.
[0055] F3: 5'-AGTACCAGTGATCATGTTCTTG-3'.
[0056] B3: 5'-TCCTGTCCACAACCTCAA-3'.
[0057] FIP: 5'-AAGTTCGAGACTCCTCGCTACTGAGGAAGTAGGATGAGGTATGA-3'.
[0058] BIP: 5'-CTCGGCCTTGAGCTTGTCAACAATAGGAAGCCGCTGAG-3'.
[0059] Loop F: 5'-TCACCGTCATTGGTATGTTGT-3'.
[0060] Loop B: 5'-AGGCGTACTTGAAGGAACC-3'.
Embodiment 2
[0062] Genomic DNA of the Fusarium pathogenic strains was extracted using the Biospin Fungal Genomic DNA Extraction Kit. The specific extraction process is as follows.
[0063] 1) Select the exfoliated Fusarium pathogenic bacteria cultured on PDA medium for 7-10 days, pick fresh mycelia, grind them into powder, and put them into a 1.5mL centrifuge tube.
[0064] 2) Add 500μL LE Buffer and mix well.
[0065] 3) Incubate at 65°C for 60 minutes (mix by inverting once every 15 minutes), and then remove.
[0066] 4) Add 130 μL DA Buffer, mix well, and bathe in water at 65°C for 5 minutes.
[0067] 5) Centrifuge at 14,000×g for 3 minutes.
[0068] 6) Take the supernatant and transfer it to a new 1.5mL centrifuge tube.
[0069] 7) Add 500μL E Binding Buffer and mix well.
[0070] 8) Transfer the mixture to the Spin Column and centrifuge at 6,000×g for 1 min.
[0071] 9) Add 500μL G Binding Buffer to the Spin Column, centrifuge at 10,000×g for 30s, and discard the liquid in the ...
Embodiment 3
[0076] Based on the LAMP primer composition designed in Example 1, the DNA extracted in Example 2 was used as a template to establish a LAMP detection reaction system for Fusarium sp.
[0077] The LAMP detection reaction system includes 0.2 μM each of the outer primers F3 and B3, 0.8 μM each of the inner primers FIP and BIP, 0.4 μM each of the loop primers Loop F and Loop B, 2×LAMP PCR isothermal amplification reaction mixture (universal type, Sangon Bioengineering (Shanghai) Co., Ltd.) 12.5μL, 8U Bst 0.5 μL of polymerase, 1.0 μL of DNA template, made up to 25 μL with sterilized ultrapure water.
[0078] Preliminarily set the reaction temperature of the LAMP reaction at 62° C. and the time for 30 minutes.
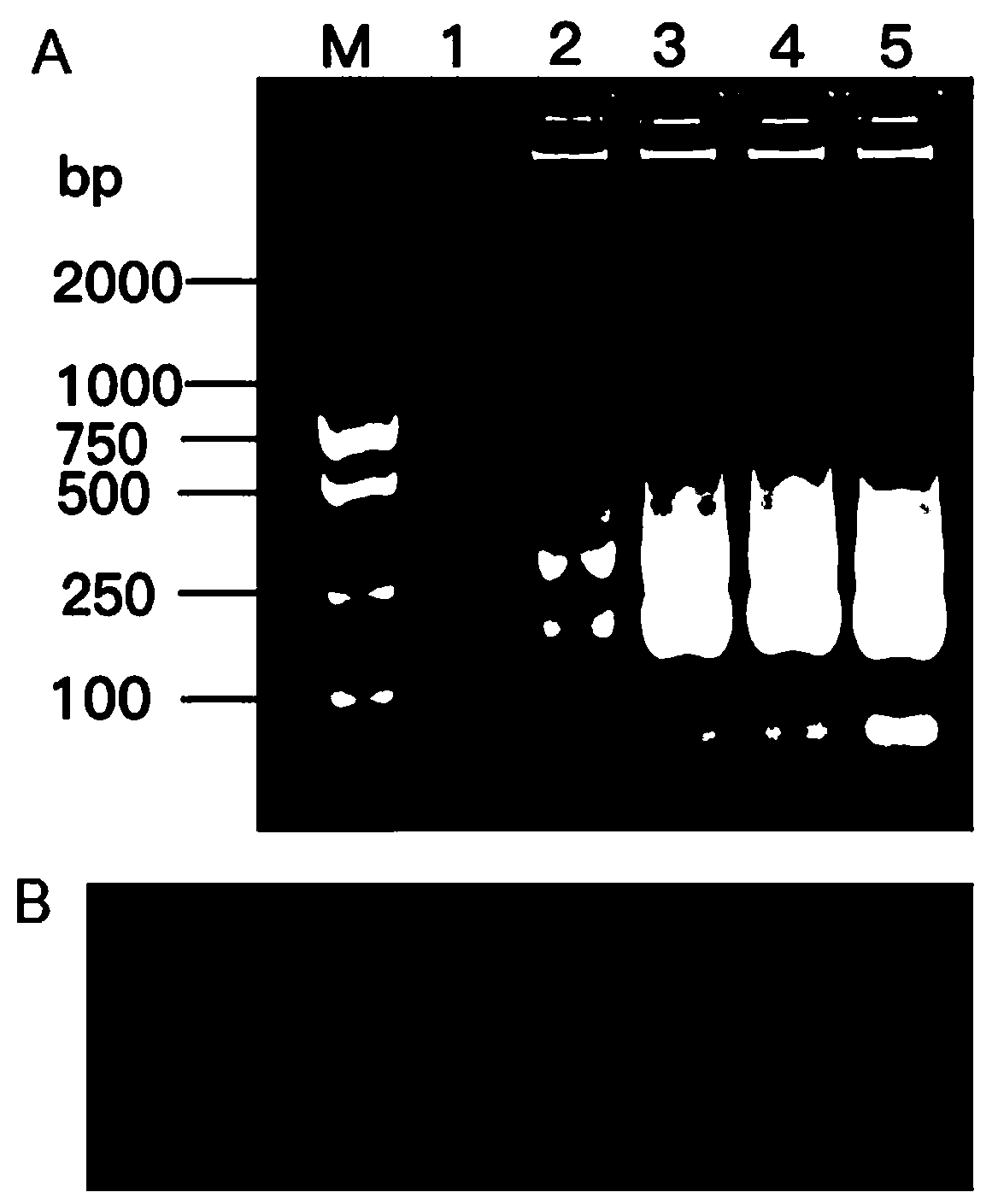
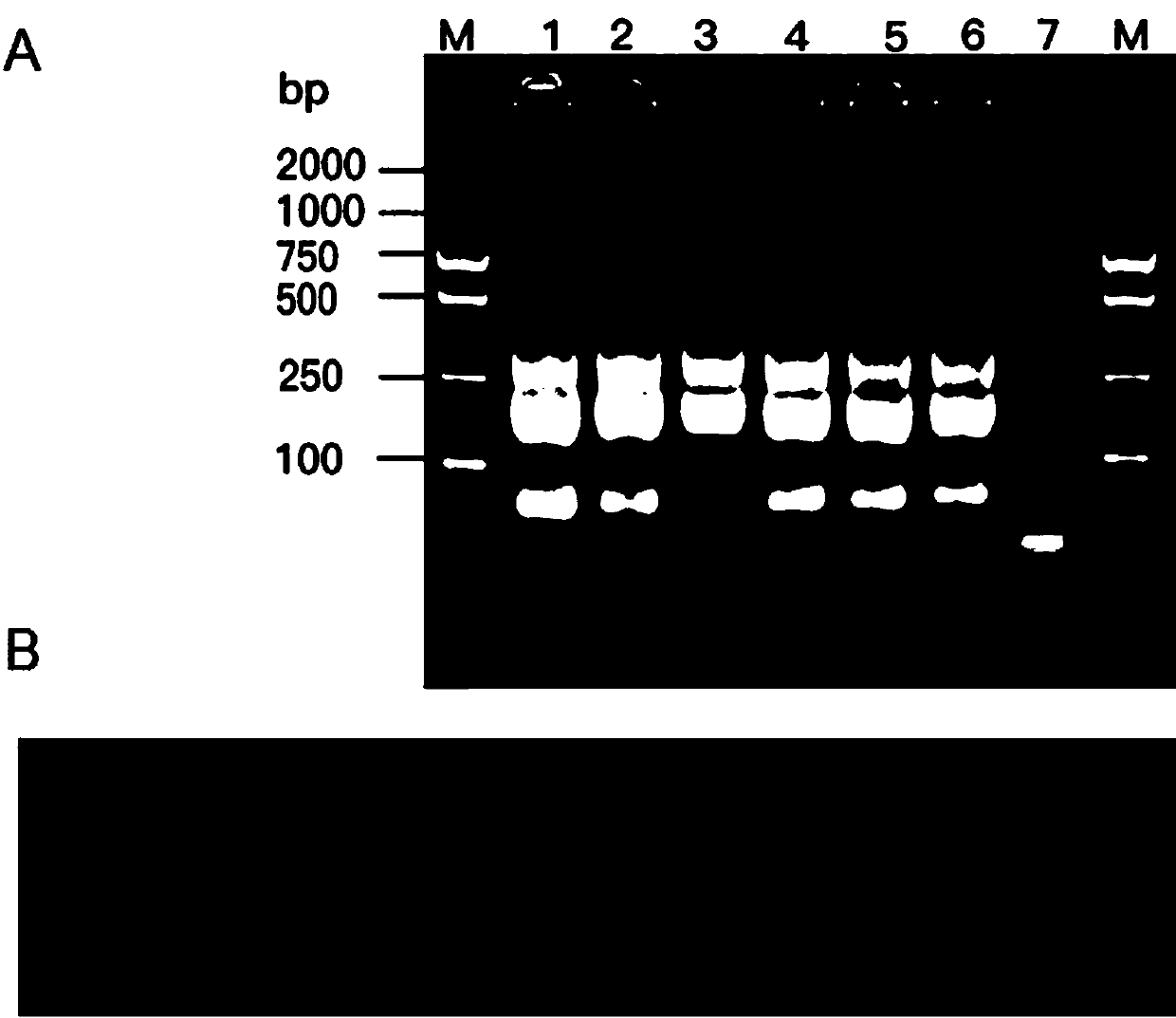
[0079] After the reaction, 2.0 μL of the LAMP amplification product was taken for 2.0% agarose gel electrophoresis detection. The electrophoresis detection conditions were voltage 80V, current 110A, and detection time 30 min.
[0080] If there is a trapezoidal band in the...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com