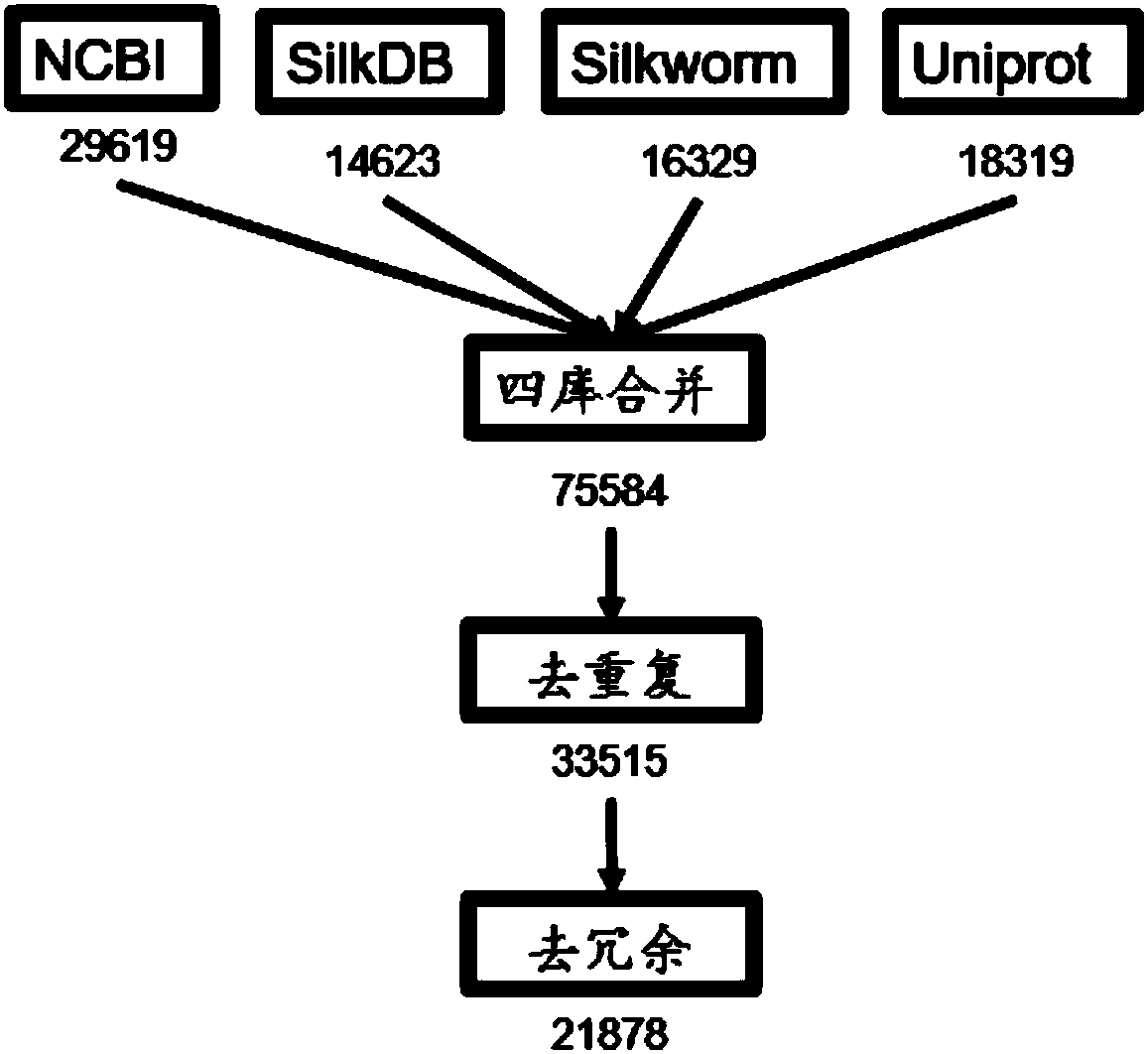
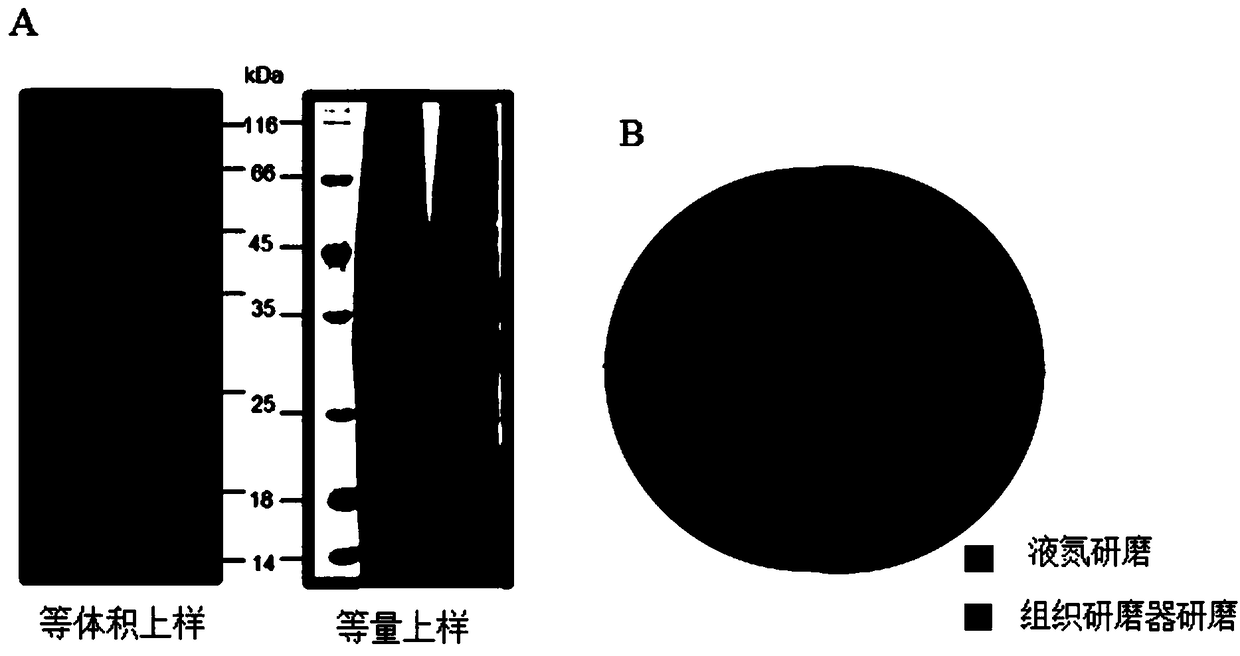
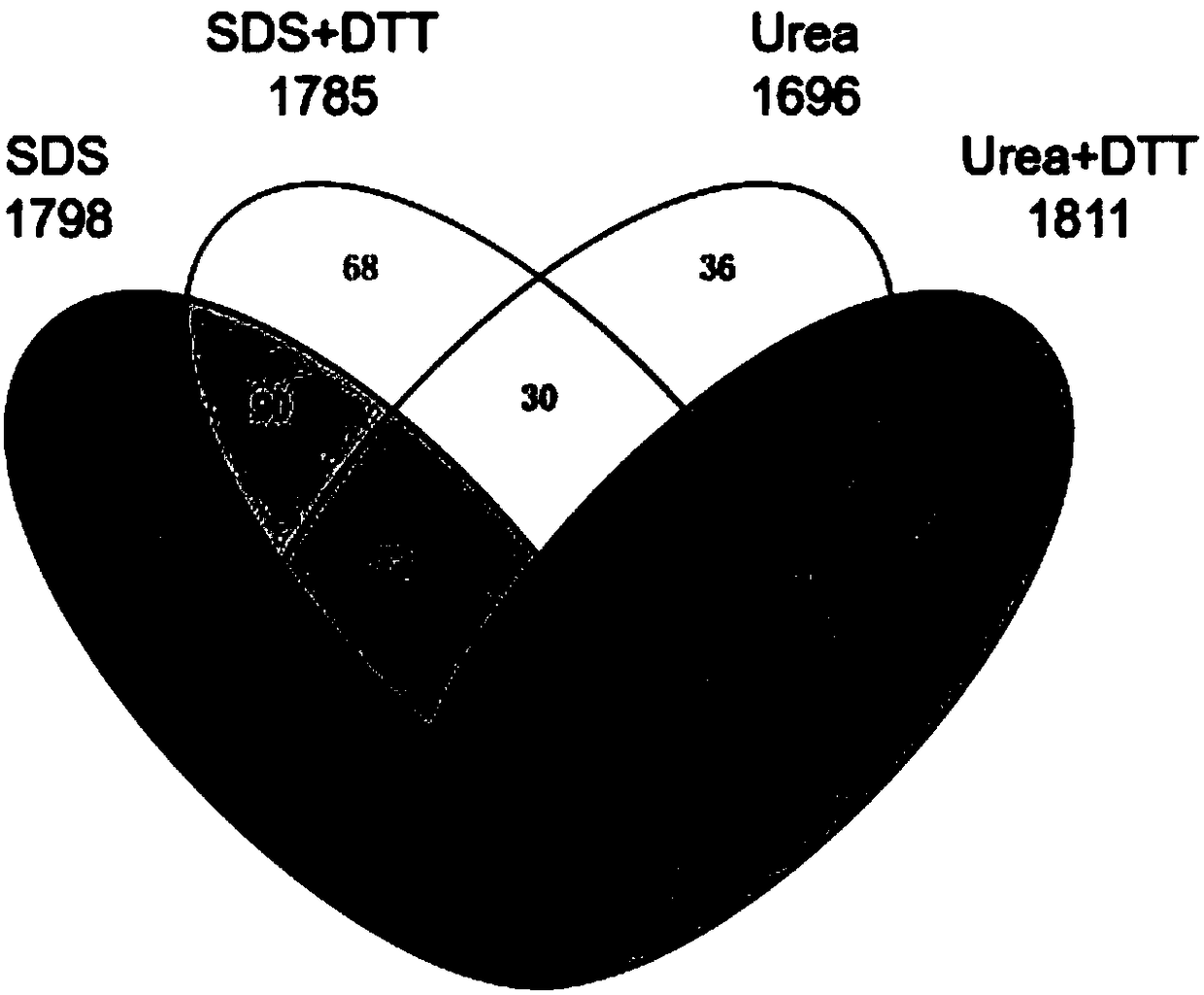
Method for carrying out large-scale proteomics identification based on silkworm tissue sample
A technology for proteomics and tissue samples, which is applied in the field of proteomics to achieve the effects of shortening sample pretreatment time, increasing the number of proteomic identifications, and reducing detection time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0053] Embodiment 1, the method for carrying out large-scale proteomic identification based on silkworm tissue sample
[0054] (1) Sample pretreatment for silkworm fat body proteomics:
[0055] a. Sample extraction
[0056] 1) Take about 2mg silkworm fat body sample and place it in a 1.5mL centrifuge tube, add 400μL 100mM DTT / 8M urea solution, put it on ice, and grind it with a Kayoudi G50 tissue grinder for 3min;
[0057] 2) Shake with a horizontal shaker for 5 minutes to fully dissolve the sample in the solution;
[0058] 3) Centrifuge at 12000g at 25°C for 15min, take the supernatant and put it in a new centrifuge tube;
[0059] 4) The protein concentration of the sample was detected with a Bradford protein quantification kit.
[0060] b. Sample pretreatment
[0061] 1) Take 300 μg of sample and add it to a 10kDa ultrafiltration tube, take 8M urea solution to make up to 200 μL, blow and aspirate several times with a pipette gun to mix evenly, and place it in a centrifug...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com