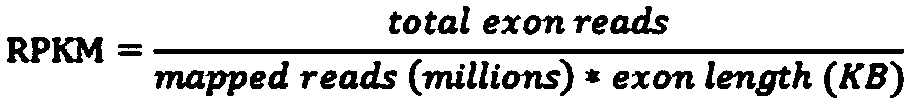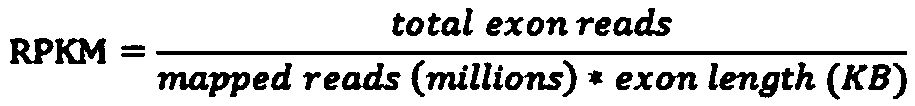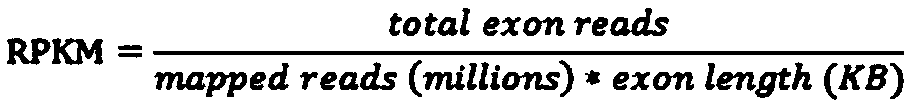Transcriptome-based tumor neoantigen identification method
A tumor antigen and identification method technology, applied in the field of bioinformatics identification of tumor neoantigens, can solve the problems of long time, low throughput and high cost, and achieve the effects of improving accuracy, reducing false negatives and shortening analysis time.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0041] A method for identifying tumor antigens based on transcriptomes, comprising the following steps:
[0042] S1: Obtain the RNA sample of the patient's tumor tissue, build and amplify the RNA sample, and obtain the sequencing result of the RNA sample of the tumor tissue.
[0043] S2: Align the short reads of the RNA sample sequencing results to the human reference genome to obtain the RNA alignment results.
[0044]S3. According to the RNA comparison result and calculate the gene expression level, perform mutation detection according to the RNA comparison result, and predict the fusion gene event according to the RNA comparison result; predict the HLA type according to the comparison result; calculate the gene expression level, mutation detection and Prediction of fusion gene events occurs in the specified order, or simultaneously.
[0045] S4: The gene expression level of the transcriptome sample; the depth of the transcriptome mutation site in the whole-external sequenc...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com