Chitin enzyme and application thereof
A technology of chitin enzyme and chitin, applied in the direction of application, enzyme, enzyme, etc., can solve the problems of low product yield, environmental burden, and large consumption of chemical reagents
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0021] Cloning of embodiment 1 chitinase gene saChiA4
[0022] The applicant used Streptomyces albolongus (ATCC 27414) to ferment chitin, and found that the amount of chitobiose in its product was significantly higher than that of other strains. Therefore, the genome of the strain was analyzed, and the chitinase obtained in the present invention was found.
[0023] According to the obtained full-length sequence of the chitinase gene, the upstream and downstream primers for amplification were designed, respectively SaChiA4F 5'-CCGGAATTCATGGAACGCGTACTACCC-3' and SaChiA4R5'-CCCAAGCTTGCAGGCGCCGTTGTCCGC-3'. The genome was used as a template for PCR amplification.
[0024] The PCR reaction system is: 2×PCR Buffer 25 μl, dNTP 5 μl, upstream and downstream primers 1 μl, template 1 μl, KOD Fx DNA polymerase (TOYOBO, KFX-101) 1 μl, add sterile water to a final volume of 50 μl.
[0025] The PCR reaction conditions were: pre-denaturation at 94°C for 2 min, denaturation at 98°C for 10 s,...
Embodiment 2
[0028] Expression of embodiment 2 recombinant protein
[0029] The correct recombinant plasmid pET28a::saChiA4 was transformed into competent cells of Escherichia coli BL21(DE3), spread evenly on LB plates with kana resistance (50ug / ml), and cultured overnight at 37°C. A single colony was picked and inserted into a test tube containing kana resistance (50ug / ml), and cultured at 37°C and 220rpm for 12h. Inoculate 1% to 2% of the inoculum into ZYP-5052 self-induction medium and induce at 20°C for 48 hours. After induction, transfer the bacterial solution to a 50mL centrifuge tube and centrifuge at 4°C for 10min to collect the cells.
[0030] ZYP-5052 medium: 1.0% peptone, 0.5% yeast powder, 0.7% Na 2 HPO4, 0.7%KH 2 PO4, 0.3% (NH4) 2 SO4, 0.05% MgSO 4 , 0.5% glycerin, 0.05% glucose, 0.2% α-lactose.
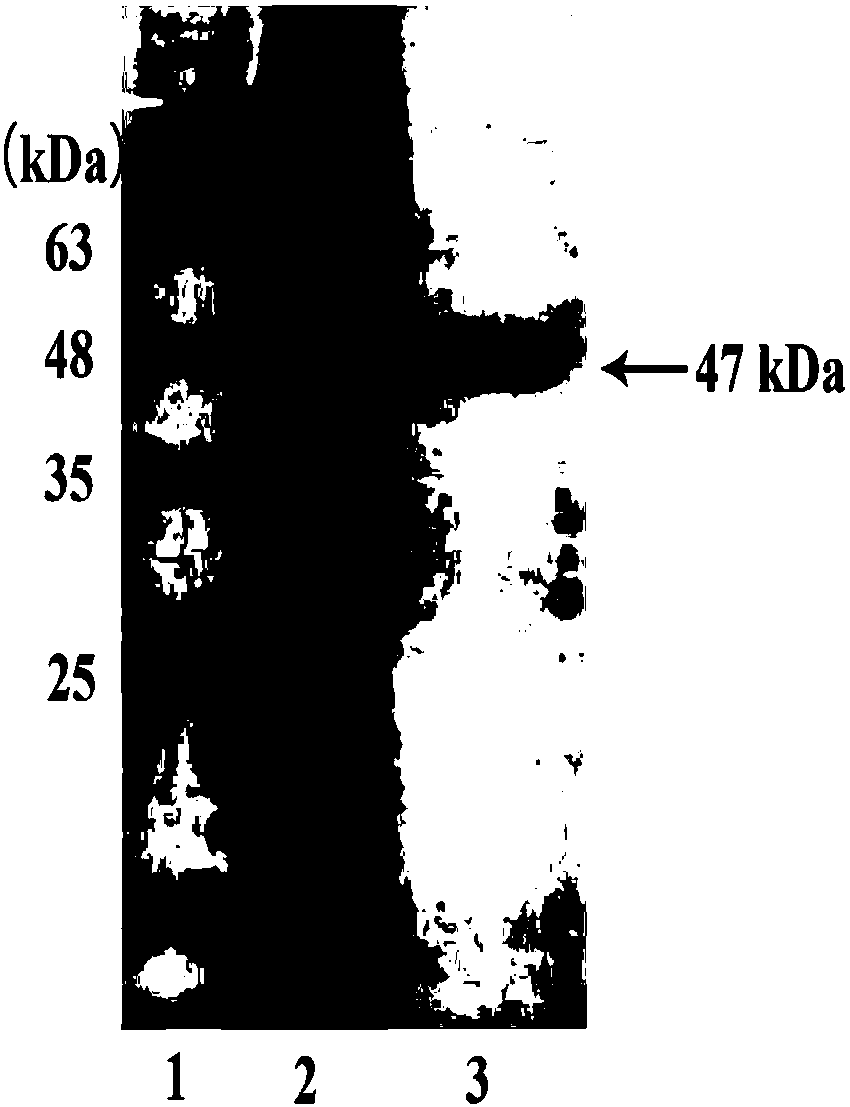
[0031] Using the principle of affinity chromatography, a Ni column was used to purify the protein of the tag expression vector containing 2 histidines. The cell pellet was firs...
Embodiment 3
[0032] The mensuration of embodiment 3 enzyme activity
[0033] The chitinase activity was determined using the DNS method.
[0034] Reaction system: 195 μL colloidal chitin (0.1g / mL), 5 μL pure enzyme (5U / mL), incubate at 55°C for 30 minutes, after the reaction, add 300 μL DNS in a boiling water bath for 10 minutes, boil for 10 minutes, and develop color. Centrifuge at 10000rpm for 5min, take 200μL supernatant, and measure OD 540 . Protein concentration was measured according to the Bradford method using bovine serum albumin (BSA) as a standard. Definition of enzyme unit: An enzyme activity unit (U) refers to the amount of enzyme required to release 1 μmol of reducing sugar per minute under the optimal reaction conditions of the enzyme.
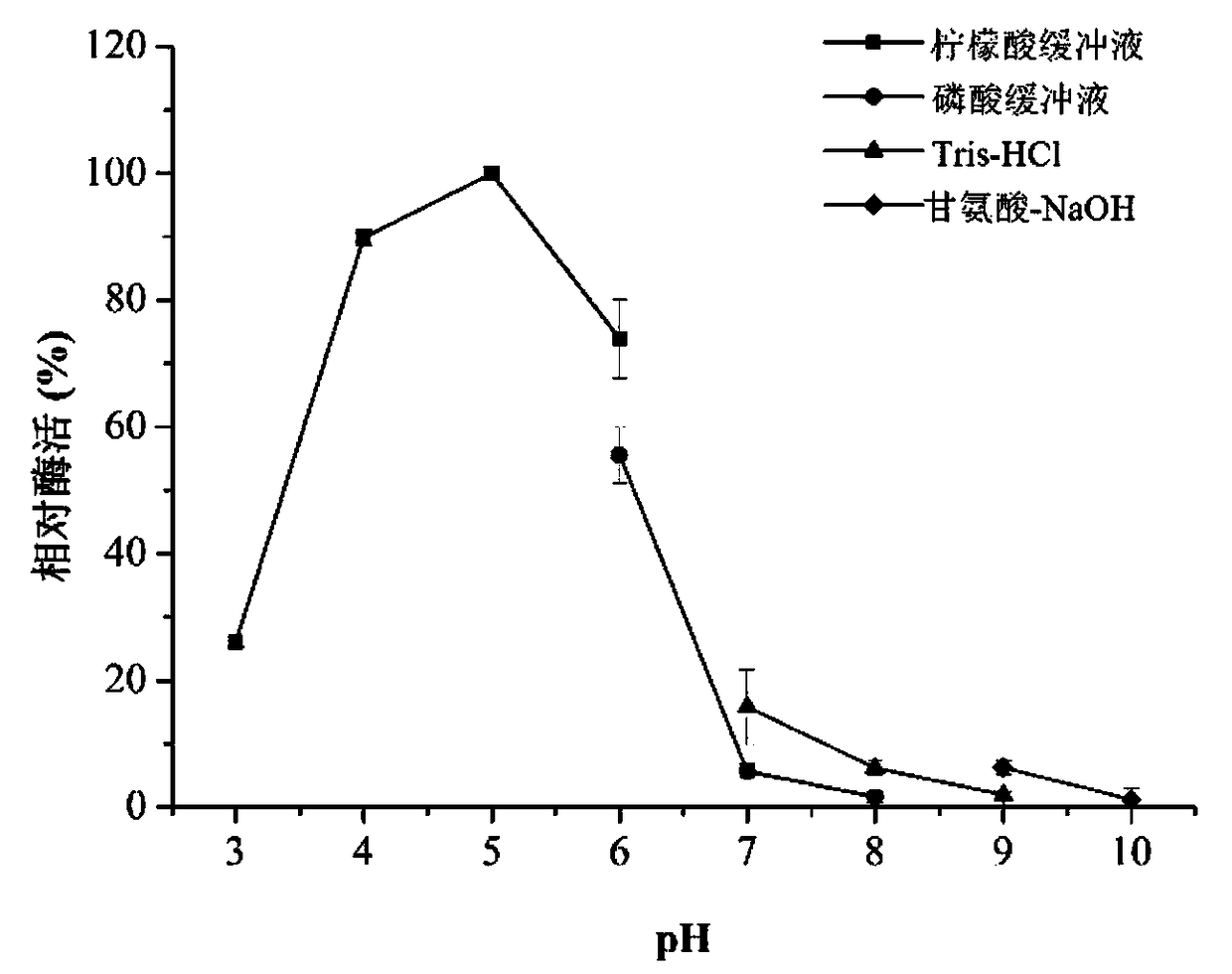
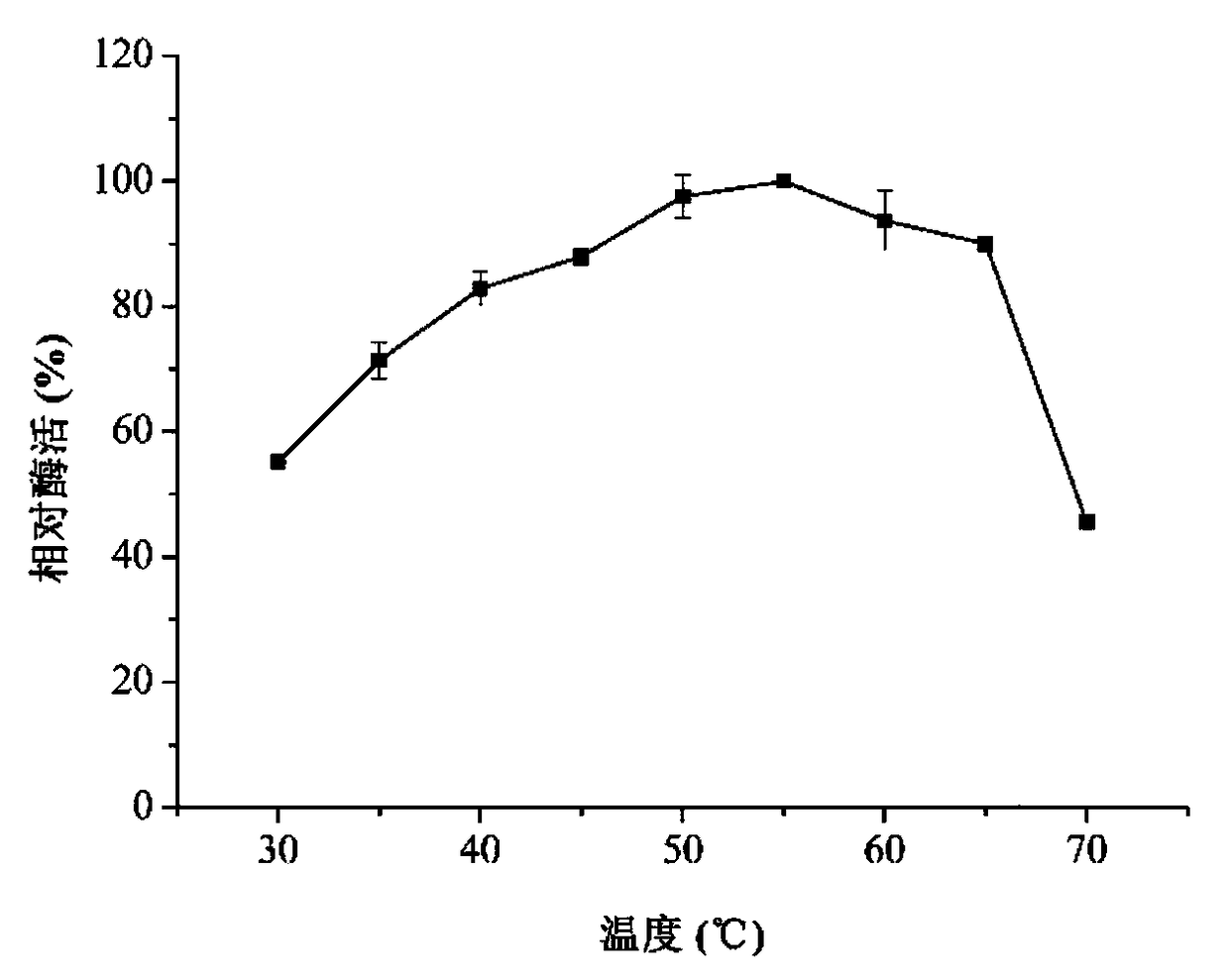
[0035] Determination of the optimal reaction conditions of chitinase.
[0036] Take an equal amount of enzyme solution (5 μL) to react at 55° C. under different pH conditions (pH 3.0-10.0), measure the enzymatic activity of chitinase, an...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com