Method for high-efficiency expression of maltogenic amylase in bacillus subtilis
A technology of Bacillus subtilis and maltogenic amylase, which is applied in the fields of genetic engineering and fermentation engineering, can solve problems such as low enzyme activity, and achieve the effects of wide sources, good expression and low production cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
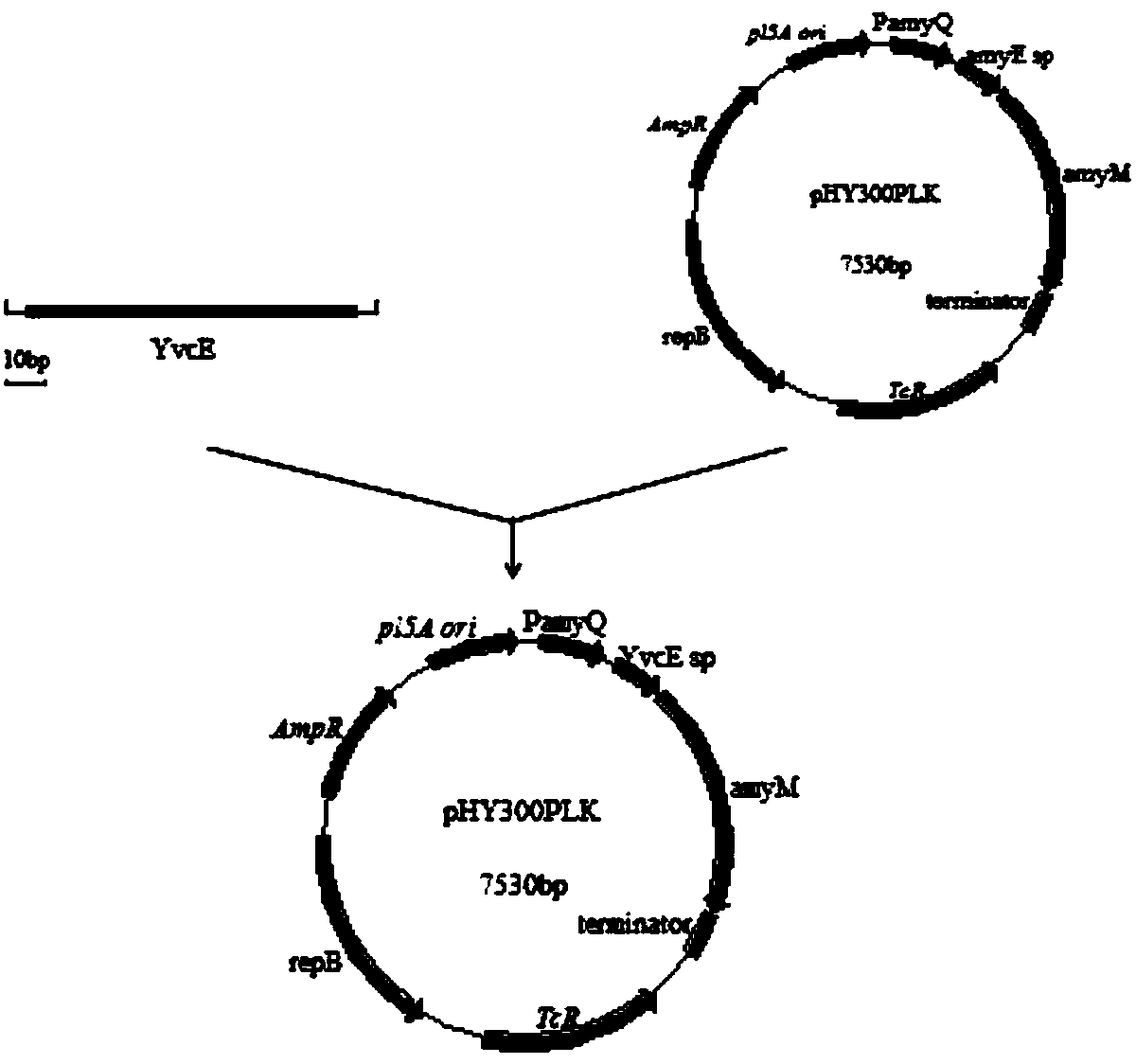
[0020] Embodiment 1: Recombinant vector construction
[0021] Recombinant construction of signal peptide YvcE, LipA, YncM and expression vector linked with maltose amylase
[0022] Primers YvcE-F and YvcE-R (SEQ ID NO.5, SEQ ID NO.6) containing homology arms were designed to amplify the YvcE signal peptide using the Bacillussubtilis168 genome as a template, and the underlined part is the region of the homology arms. The recombinant construction of LipA and YncM signal peptide is the same as above, and the primers are LipA-F, LipA-R (SEQ ID NO.7, SEQ ID NO.8) and YncM-F, YncM-R (SEQ ID NO.9, SEQ ID NO. NO.10).
[0023] Primers P-F, P-R (SEQ ID NO.11, SEQ ID NO.12) were designed to amplify the expression vector sequence using the amyM-pHY300PLK vector plasmid as a template.
[0024] YvcE-F: 5'- TAAGGAGTGTCAAGA ATGAGAAAGAGTTTAATTACACTTGGT-3'
[0025] YvcE-R: 5'- GCTTGCAGAAGAAGA CGCCGATGCAGTTTTACTTGTAAA-3'
[0026] LipA-F: 5'- TAAGGAGTGTCAAGA ATGAAATTTGTAAAAAGAAGGATCAT-3...
Embodiment 2
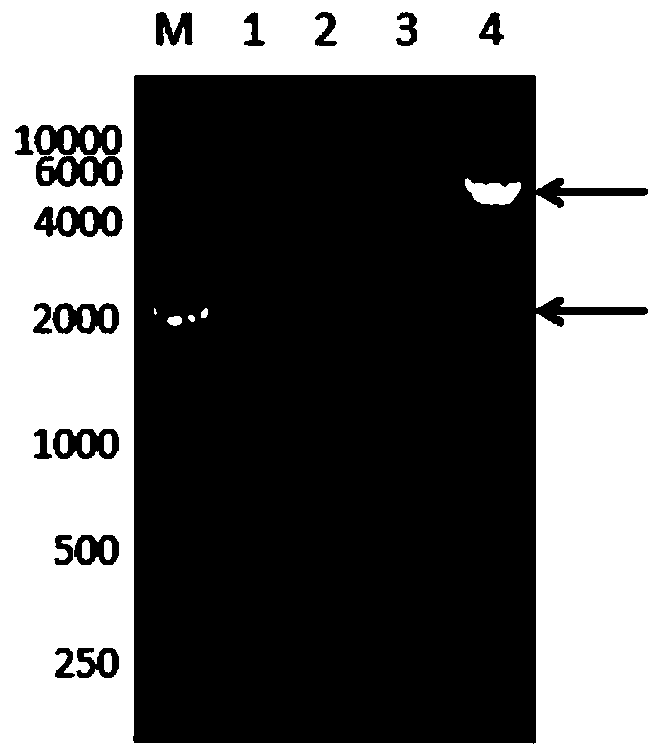
[0037] Embodiment 2: Transformation of recombinant plasmid
[0038] 1) Fresh LB plate (LB solid medium: peptone 10g / L, yeast extract 5, NaCl 10g / L, 0.2g / L agar powder) pick a single colony of Bacillus subtilis (CCTCC M2016536) and inoculate it in 5ml LB liquid culture medium, 37°C, 200rpm for 10.5h.
[0039] 2) Take 2.5 mL and transfer it into 40 mL of LB medium with 0.5 M sorbitol, culture at 37° C. with shaking at 200 rpm for 4.5 hours.
[0040] 3) Take all the bacteria liquid and bathe in ice water for 10 minutes, then centrifuge at 5000 rpm and 4°C for 5 minutes to collect the bacteria.
[0041] 4) Wash the cells with 40ml of pre-cooled electroporation buffer (sorbitol 0.5M, mannitol 0.5M, glucose 10%), centrifuge at 5000rpm, 4°C for 5min to remove the supernatant, and rinse 4 times in this way.
[0042] 5) Resuspend the washed bacteria in 1mL electroporation medium, aliquot them into 1.5mL EP tubes, and fill each tube with 300ul competent cells.
[0043]6) Add 10 μL of...
Embodiment 3
[0045] Embodiment 3: Shake flask fermentation produces enzyme and the mensuration of maltose amylase enzyme activity
[0046] The recombinant Bacillus subtilis strain obtained in Example 2 was inoculated in LB medium, and after culturing at 37°C for 8-10 hours, it was transferred to TB fermentation medium with a 5% inoculation amount, and cultivated at 37°C and 200rpm for 2h, Then move to 33 ℃ constant temperature culture for 48h to produce enzyme. After the fermentation, the supernatant collected by centrifugation is the crude enzyme liquid.
[0047] Mix 1mL of 1% soluble starch solution and 0.9mL of 50mM, pH5.5 phosphate buffer well, preheat at 60°C for 10min, add 0.1mL of enzyme solution, oscillate and mix well, add 3mL of DNS after 10min of reaction, oscillate , boiled for 7 minutes, cooled rapidly, added distilled water to make up to 15ml, and measured the absorbance at 540nm (using the inactivated enzyme solution as a catalyst, the same operation was used as a blank). ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com