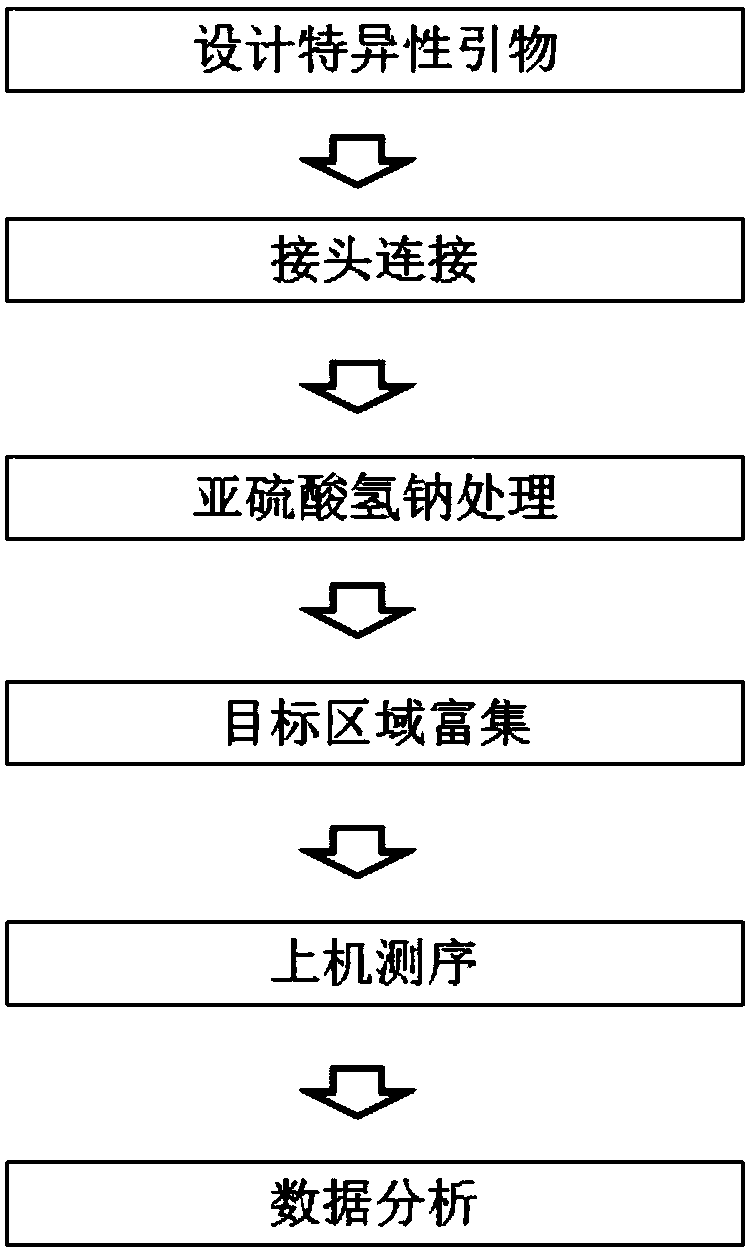
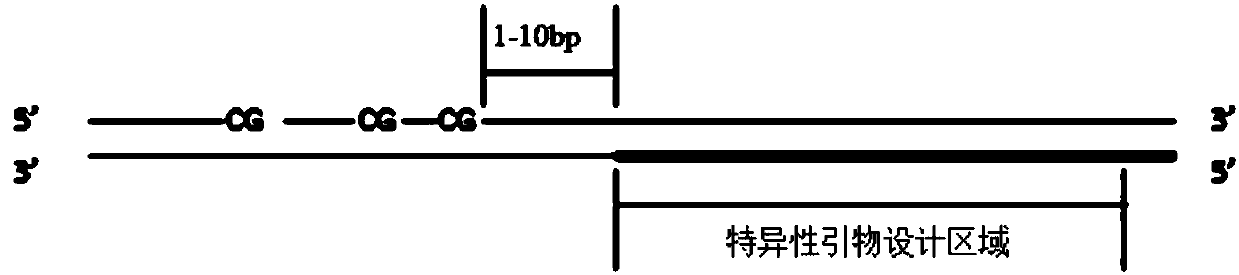
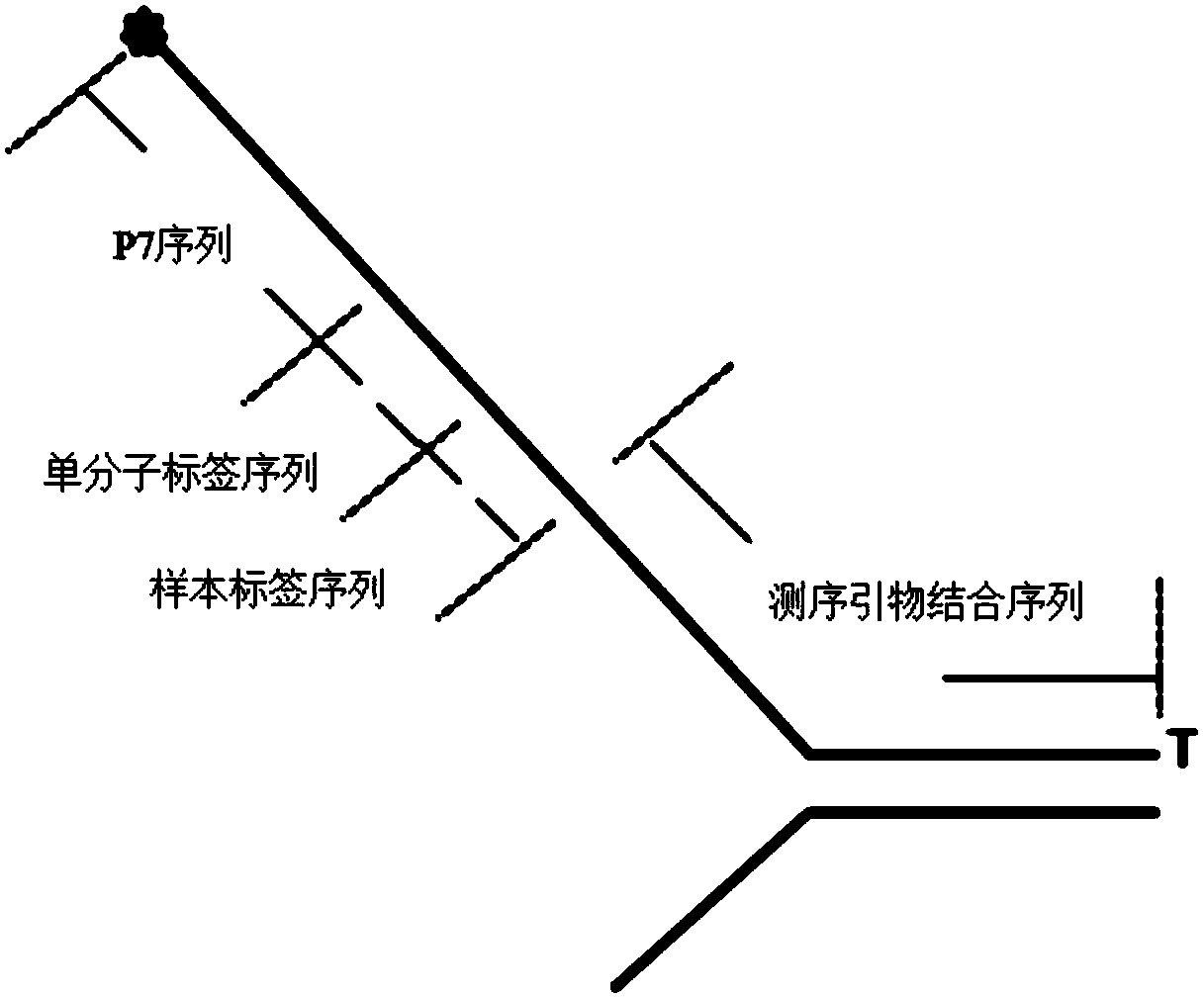
Construction method and detection method of trace fragmented DNA methylation detection library
A construction method and methylation technology, which can be used in chemical libraries, biochemical equipment and methods, and microbial determination/inspection. It can solve problems such as poor targeting, high sequencing costs, and inability to detect trace DNA.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0042] The region to be detected in this embodiment is located near the promoter region or the first exon region of the SEPT9 gene. The specific sequence is as follows: AGCCAGGGGGCCTAGGGGCTCCTCCGGCGGCTAGCTCTGCACTGCAGGAGCGCGGGCGCGGCGCCCCAGCCAGCGCGCAGGGCCCGGGCCCCGCCGGGGGCGCTTCCTCGCCGCTGCCCTCCGCGCGACCCGCTGCCCACCAGCCATCAT GTCGGACCCCGCGGTCAACGCG CAGCTGGATGGGATCATTTCGGACTTCGAAGGTGGGTGCTGGGCTGGCTGCTGCGGCCGCGGACGTGCTGGAGAGGACCCTGCGGGTGGGCCTGGCGCGGGACGGGGGTGCGCTGAGGGGAGACGGGAGTGCGCTGAGGGGAGACGGGAC (SEQ ID NO: 1).
[0043] 1. Design specific primers for the above regions:
[0044] SPT9-S:
[0045] ACACTCTTTCCCTACACGACGCTCTTCCGATCTTTCGAAATCCGAAATAATCCCATCCAACTA (SEQ ID NO: 2).
[0046] 2. Design and manufacture the linker sequence of the illumina sequencing platform:
[0047] ADT-S: GATCGGAAGAGC (SEQ ID NO: 3).
[0048] All cytosine Cs in the linker sequence are methylated.
[0049] ADT-AS:
[0050] CAAGCAGAAGACGGCATACGAGATNNNNNNNIIIIIIIGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT (SEQ ID N...
Embodiment 2
[0096] 1. The specific sequence of the area to be detected in this embodiment is as follows:
[0097]CpG island region of SEPT9 gene:
[0098] AGCCAGGGGGCCTAGGGGCTCCTCCGGCGGCTAGCTCTGCACTGCAGGAGCGCGGGCGCGGCGCCCCAGCCAGCGCGCAGGGCCCGGGCCCCGCCGGGGGCGCTTCCTCGCCGCTGCCCTCCGCGCGACCCGCTGCCCACCAGCCATCATGTCGGACCCCGCGGTCAACGCGCAGCTGGATGGGATCATTTCGGACTTCGAAGGTGGGTGCTGGGCTGGCTGCTGCGGCCGCGGACGTGCTGGAGAGGACCCTGCGGGTGGGCCTGGCGCGGGACGGGGGTGCGCTGAGGGGAGACGGGAGTGCGCTGAGGGGAGACGGGAC(SEQ ID NO:7)。
[0099] CpG island region of NDRG4 gene:
[0100] TCTAAGGTTCCCCTTGGGAGTCTAAACAAAGACTACGGCAGCGCCGTCCCCTCCCCCGGGAACCCGACGCCGCGCGGCCACAGGGGGCCTGGAGGGGCGGGCAGGGCCTCGCAGCGCACCCAGCACAGTCCGCGCGGCGGAGCGGGTGAGAAGTCGGCGGGGGCGCGGATCGACCGGGGTGTCCCCCAGGCTCCGCGTCGCGGTCCCCGCTCGCCCTCCCGCCCGCCCACCGGGCACCCCAGCCGCGCAGAAGGCGGAAGCCACGCGCGAGGGACCGCGGTCCGTCCGGGACTAGCCCCAGGCCCGGCACCGCCCCGCGGGCCGAGCGCCCACACCCGCCAAACCCACGCGGGCACGCCCCCGCGGCGCACCGCCCCCAGCCCGGCCTCCGCCCCTGCAGCCGCGGGCACGCGGAGGGGCTCCTGGCTGCCCGCACCTGCACCCGCGCGTCGGCGGC(SE...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com