Real-time quantitative PCR detection primer set and method of differentially expressed acipenser dabryanus gonad gene
A differentially expressed gene, real-time quantitative technology, applied in the field of molecular biology, can solve the problem of not analyzing the differential gene expression of sturgeon gonads
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0027] The selection of embodiment 1 object detection gene and internal reference gene:
[0028] 1. Dabry's sturgeon sample collection and RNA extraction:
[0029] Select healthy, gonad-differentiated Acipenser dabryi from the Sichuan Fisheries Research Institute, dissect their gonads to identify their sex, and extract the remaining samples for total RNA. In a pre-cooled mortar, use liquid to quickly grind into powder, put it into a 1.5mL centrifuge tube, and add 1mL TRIzol; (2) Add 200μL chloroform to the centrifuge tube, shake vigorously for 15s, and stand at room temperature for 3min; (3) 4°C , centrifuge at 12,000r / min for 10min, absorb the supernatant and put it into a new centrifuge tube; (4) add isopropanol equal to the volume of the supernatant, shake gently to mix, and place at room temperature for 5-10min; (5) 4°C, Centrifuge at 12,000r / min for 10min, discard the supernatant, and carefully absorb the remaining isopropanol; (6) Add 1mL of 75% ethanol to wash the prec...
Embodiment 2
[0038] Example 2 Establishment of real-time quantitative PCR detection method for differentially expressed genes in gonads of Acipenser dabfieldii:
[0039] 1. Primer design:
[0040]According to the sequences of SOX9, FST and DF in the transcriptome unigene gene, Primei5.0 software was used to design specific primers suitable for fluorescent quantitative PCR detection. The internal reference gene β-actin also designed primers according to the transcriptome unigene results. The primers are shown in Table 1. Shown:
[0041] Table 1 Specific primers suitable for fluorescent quantitative PCR detection
[0042]
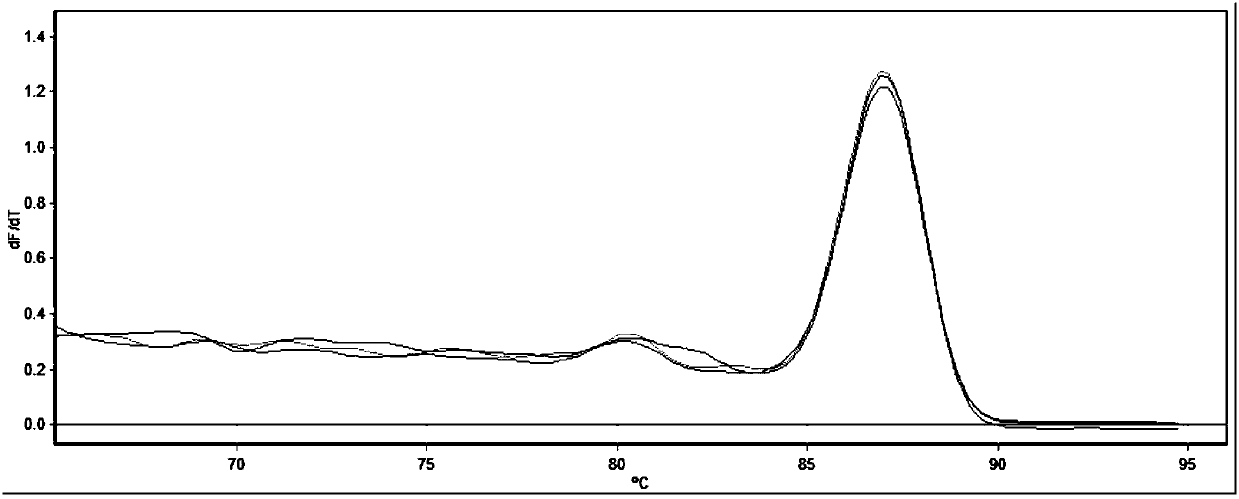
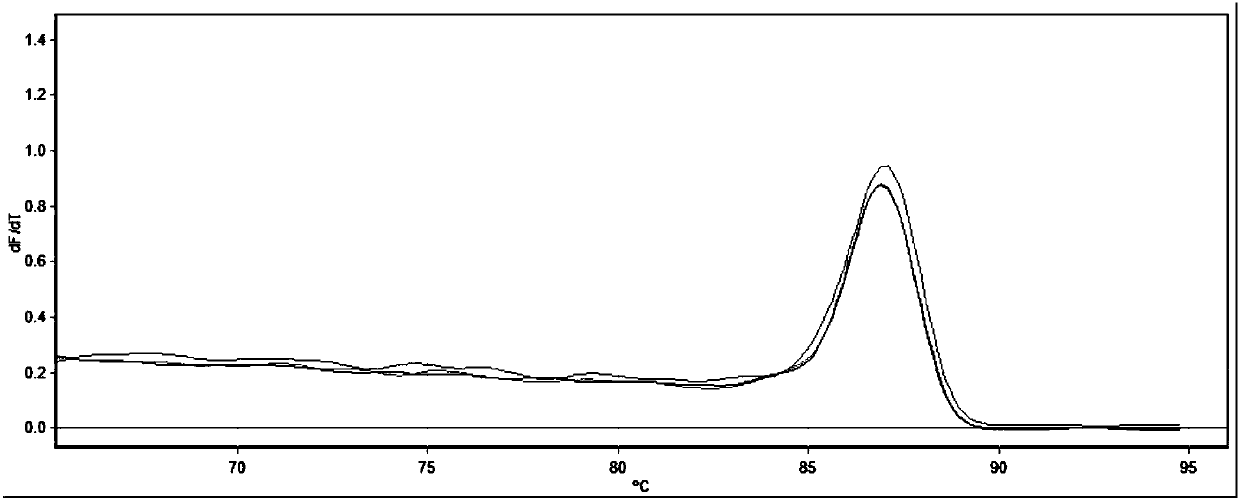
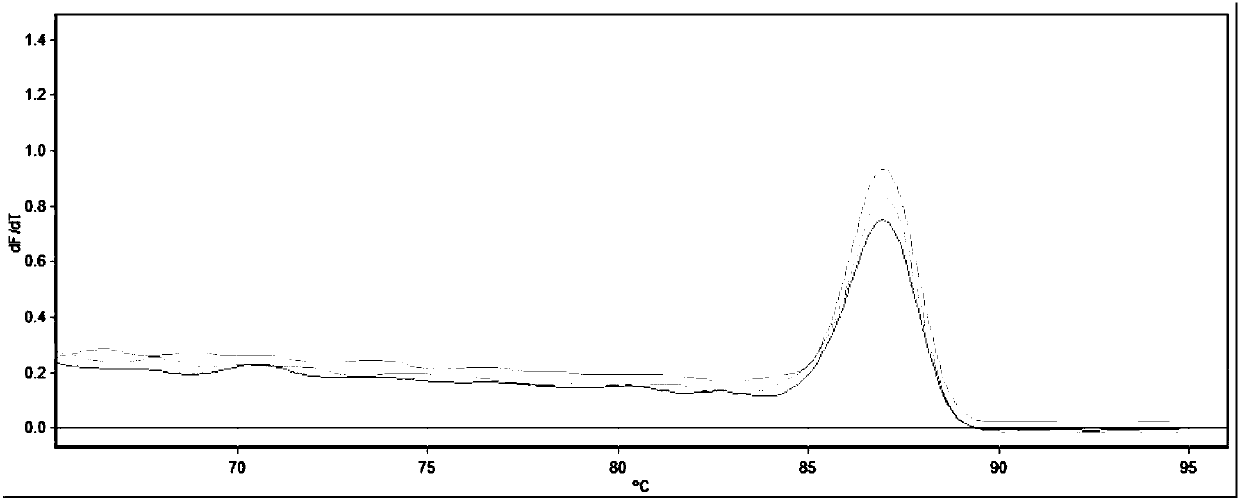
[0043] The result of agarose gel electrophoresis shows that the amplified length is consistent with the expected product length, and the sequence is determined to be the detected target gene (shown in SEQ ID NO.1-4), as shown in Figure 1-4 shown, from figure 1 It can be seen that the melting curve of the SOX9 gene is a single peak, indicating that the primer has go...
Embodiment 3
[0055] Embodiment 3 quantitative PCR result statistics:
[0056] Table 2 below compares the expression levels of the three genes SOX9, DF and FST in quantitative PCR and transcriptome sequencing.
[0057] Calculated as follows:
[0058] ΔCt=Ct(target gene)-Ct(β-actin gene)
[0059] ΔΔCt = ΔCt (male sample) - ΔCt (female sample)
[0060] to 2 -ΔΔCt Values represent relative expression levels of target genes.
[0061] Table 2 Comparison of transcriptome sequencing and quantitative PCR results
[0062]
[0063] FPKM is the expression level of transcriptome sequencing, qPCR is the expression level of quantitative PCR; T: testis; O: ovary.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com