CRISPR-mediated rapid and effective crop site-specific gene fragment or allele replacement method and system
An allele and fragment technology, applied in the field of allele replacement, can solve the problems of time-consuming and laborious, low HDR frequency, etc., and achieve the effect of great application potential and application prospect.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0050] Example 1. Preparation of recombinant plasmid pCXUN-cas9-gRNA1-gRNA2-arm donor and free donor fragment
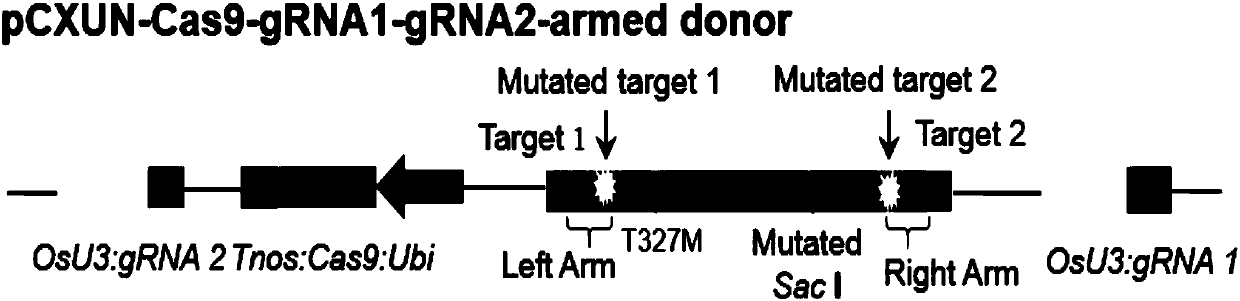
[0051] The free donor fragment is shown in sequence 4 of the sequence listing. The schematic diagram of the structure of the free donor fragment is shown in figure 1 shown. In sequence 4 of the sequence listing, the 4th-26th nucleotide is target 1 (Target1) [the 4th-23rd nucleotide is the target sequence of sgRNA1, and the 24th-26th nucleotide is CGG], the 30th -The 129th nucleotide is the upstream homology arm (Left Arm), the 130th-152nd nucleotide position is mutated target 1 (Mutated target1) [the 130th-149th nucleotide is the target sequence of sgRNA1, the 150th -The 152nd nucleotide is CCG], the 155th-157th nucleotide is ATG (coding methionine) [that is, causes the T327M mutation in the protein], the 296th-301st nucleotide is GAACTC (Mutated SacⅠ )[is about to mutate the original restriction endonuclease SacⅠ "GAGCTC"], the 358th-380th nucleotide is the mutat...
Embodiment 2
[0054] Example 2, Application of recombinant plasmid pCXUN-cas9-gRNA1-gRNA2-arm donor and free donor fragments to introduce site-directed mutations in the NRT1.1B gene
[0055] 1. Obtaining genetically modified rice
[0056] 1. Take the plump 11 seeds of Zhonghua, peel off the seed coat, sterilize and wash in sequence, then place on the induction medium, and culture in the dark at 28°C for 40-50 days to induce the generation of callus.
[0057] Induction medium: solid NB medium containing 2mg / L 2,4-D.
[0058] 2. After completing step 1, take the callus and treat it on the hyperosmotic medium for 4-6 hours,
[0059] Hypertonic medium: solid NB medium containing 0.3M mannitol and 0.3M sorbitol.
[0060] 3. Mix the recombinant plasmid pCXUN-cas9-gRNA1-gRNA2-arm donor and the free donor fragment at a molar ratio of 1:20, and then bombard the callus that completed step 2 with a gene gun (using 0.6 μm gold powder, the bombardment pressure is 900psi).
[0061] 4. Take the callus...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com