Breast cancer marker
A marker, breast cancer technology, applied in the field of biomedicine, can solve the problems of breast cancer cell loss, loose cell connection, easy to fall off, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
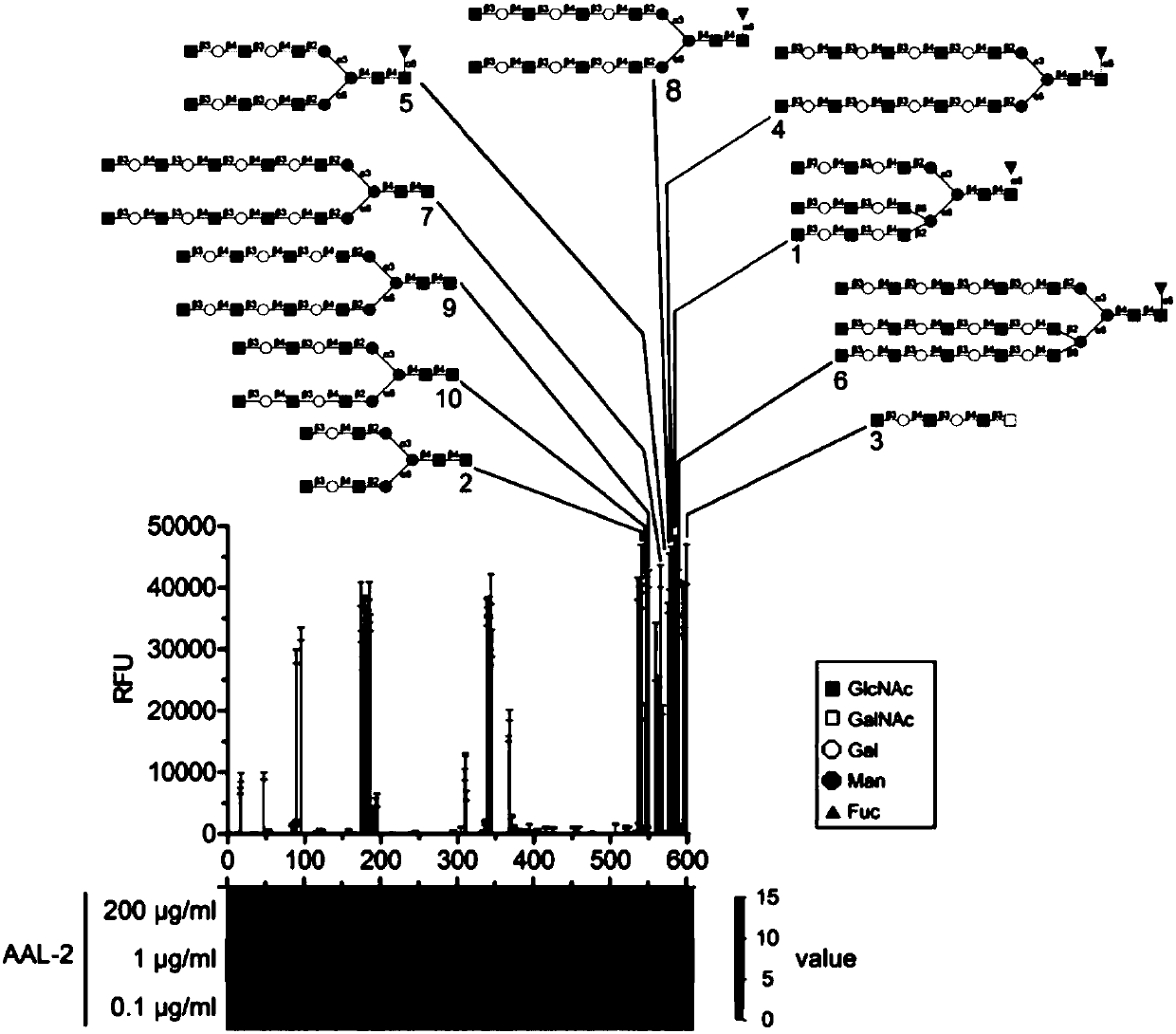
[0051] Using lectin affinity chromatography combined with high performance liquid chromatography / mass spectrometry to identify differentially expressed glycoproteins in the plasma of breast cancer patients in order to obtain breast cancer candidate markers.
[0052] 1. Sample source and information
[0053]Plasma samples were collected from Dongguan Sixth People's Hospital and Zhongshan Xiaolan Hospital. According to the seventh edition of the American Joint Commission for Cancer Staging (AJCC) clinical staging criteria for breast cancer, they were divided into Stage0: TisN0M0; StageⅠ: T1N0M0; StageⅡA: T0N1M0, T1N1M0, T2N0M0; StageⅡB: T2N1M0, T3N0M0; StageⅢA: T0N2M0, T1N2M0, T2N2M0, T3N1-2M0; StageⅢB: T4N0-2M0; StageⅢC: any T N3M0; StageⅣ: any T any N M1stage. Among them, Tis is carcinoma in situ, T stands for primary tumor, N stands for regional lymph nodes, and M stands for distant metastasis. Breast cancer samples are grouped here according to their sex, age and number of ...
Embodiment 2
[0070] The above candidate markers were further verified by western blot.
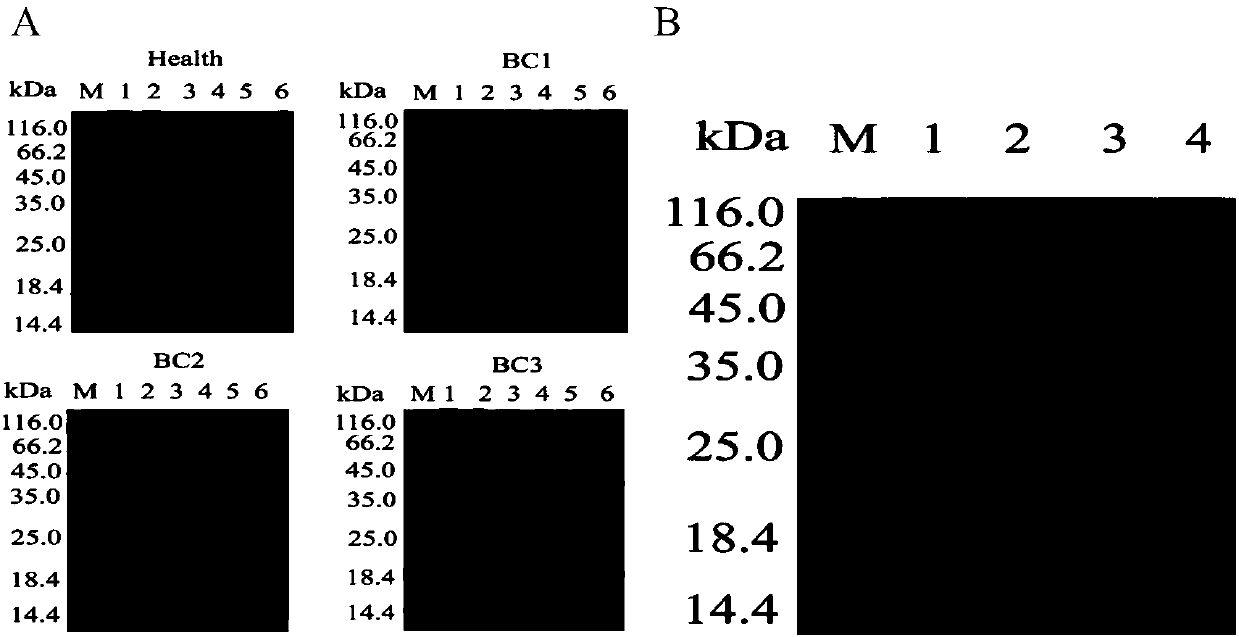
[0071] In order to further verify the expression changes of these three proteins, the inventors used W estern blo t The protein expression changes in 52 plasma samples of 19 cases of BC1, 9 cases of BC2, 8 cases of BC3, 4 cases of BC4 and 12 healthy persons were detected. For each set of Wes tern blo t In the experiment, healthy control H1 was selected as a standard, and other bands were quantified using WB grayscale analysis software, and each protein band was quantified by densitometry. The result is as Figure 6 As shown, Serotransferrin protein expression was down-regulated in Stage1 group compared with healthy control group (consistent with mass spectrometry data); Plasma protease C1 inhibitor was significantly down-regulated in Stage4 group compared with healthy control group (inconsistent with mass spectrometry data); SerpinB4 Compared with the healthy control group and Stage1 group, it wa...
Embodiment 3
[0074] The above candidate markers were further verified by reverse lectin ELISA (Reverse-Lectin ELISA). The following is the Reverse-Lectin ELISA test procedure.
[0075] Prepare 4 μg / ml of AAL2 in 15 mmol / l sodium carbonate buffer solution (pH 9.6), coat ELISA 96-well plate, 100 μl per well, overnight at 4°C; the next day, discard the coating solution and wash once with PBST; wash once with 5 % Skimmed milk powder blocking solution (5% skimmed milk powder dissolved in PBST solution, pH 7.4) 300 μl per well for blocking incubation, overnight at 4°C; discard the blocking solution, add 300 μl washing solution to each well, wash five times, 3 min each time, Pat the plate dry on filter paper; PBST dilutes the appropriate multiple of plasma samples (according to the pre-experimental dilution results), add 100 μl to each well, incubate at room temperature for 1 h; discard the samples, add 300 μl PBST washing solution to each well, wash five times, each Pat the plate dry on filter ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com