Method for obtaining bar code area of COI (C oxidase I) gene of insect in batched and high-accuracy ways by using PacBio monomolecule sequencing
A barcode and insect technology, applied in biochemical equipment and methods, microbial measurement/inspection, etc., can solve the problems of not being able to obtain the full length of the barcode area, affecting data integrity, and failing to meet the requirements of species identification, and achieve major applications The effect of promoting value, reducing time and labor costs, and improving efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0050] Embodiment 1, the preparation of primer
[0051] The nucleotide sequences of primer LCO1490 and primer HCO2198 are as follows:
[0052] LCO1490 (sequence 1 of the sequence listing): 5'-GGTCAACAAATCATAAAGATATTGG-3';
[0053] HCO2198 (sequence 2 of the sequence listing): 5'-TAAACTTCAGGGTGACCAAAAAAATCA-3';
[0054] The target sequence is 658bp.
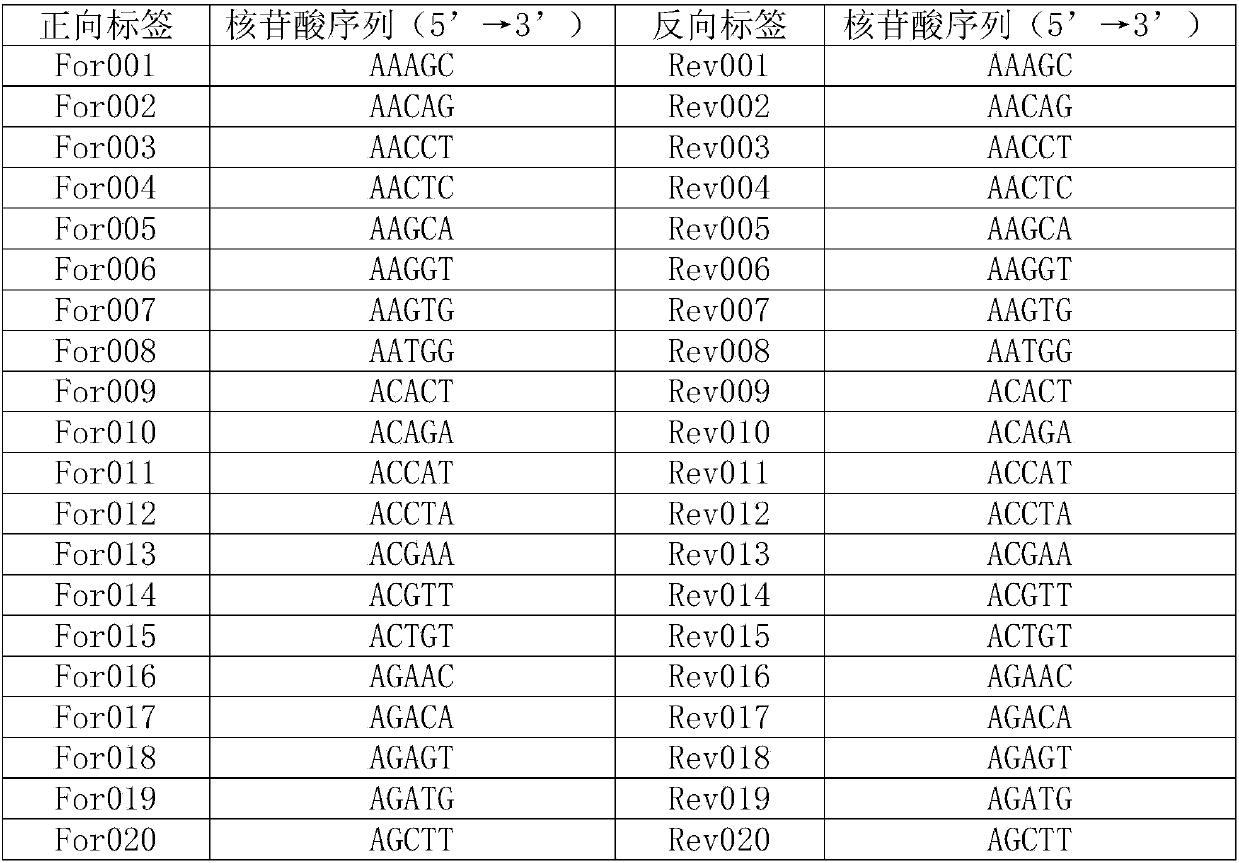
[0055] A forward tag was added to the 5' end of primer LCO1490, and a reverse tag was added to the 5' end of primer HCO2198. The nucleotide sequences of the forward tag and the reverse tag are shown in Table 1. For example, add forward label For001 to the 5' end of primer LCO1490 to obtain primer For001 (forward primer); add reverse label Rev001 to the 5' end of primer HCO2198 to obtain primer Rev001 (reverse primer); primers For001 and Primer Rev001 constitutes primer pair 001. By analogy, 96 primer pairs were obtained, sequentially from primer pair 001 to primer pair 096.
[0056] Table 1
[0057]
[0058]
[0059] ...
Embodiment 2
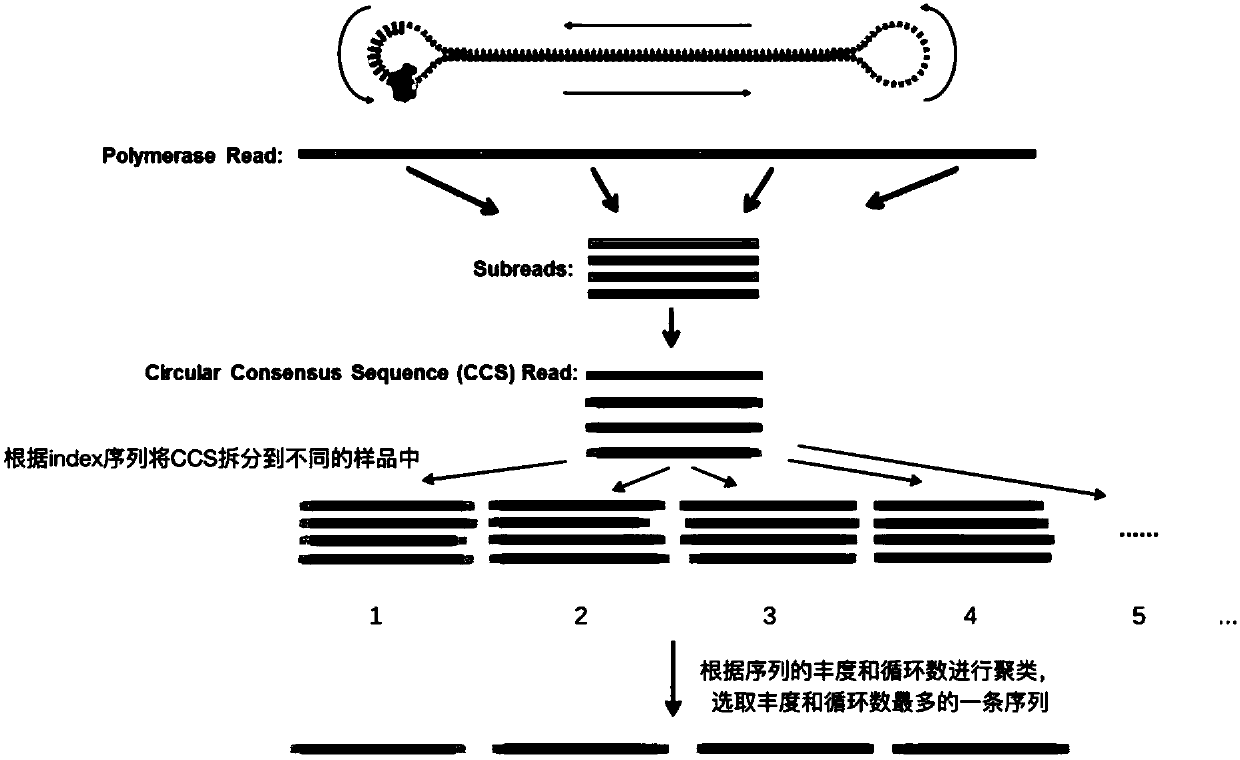
[0060] Example 2, the insect COI sequence obtained by applying the primers of Example 1 and the third generation sequencing
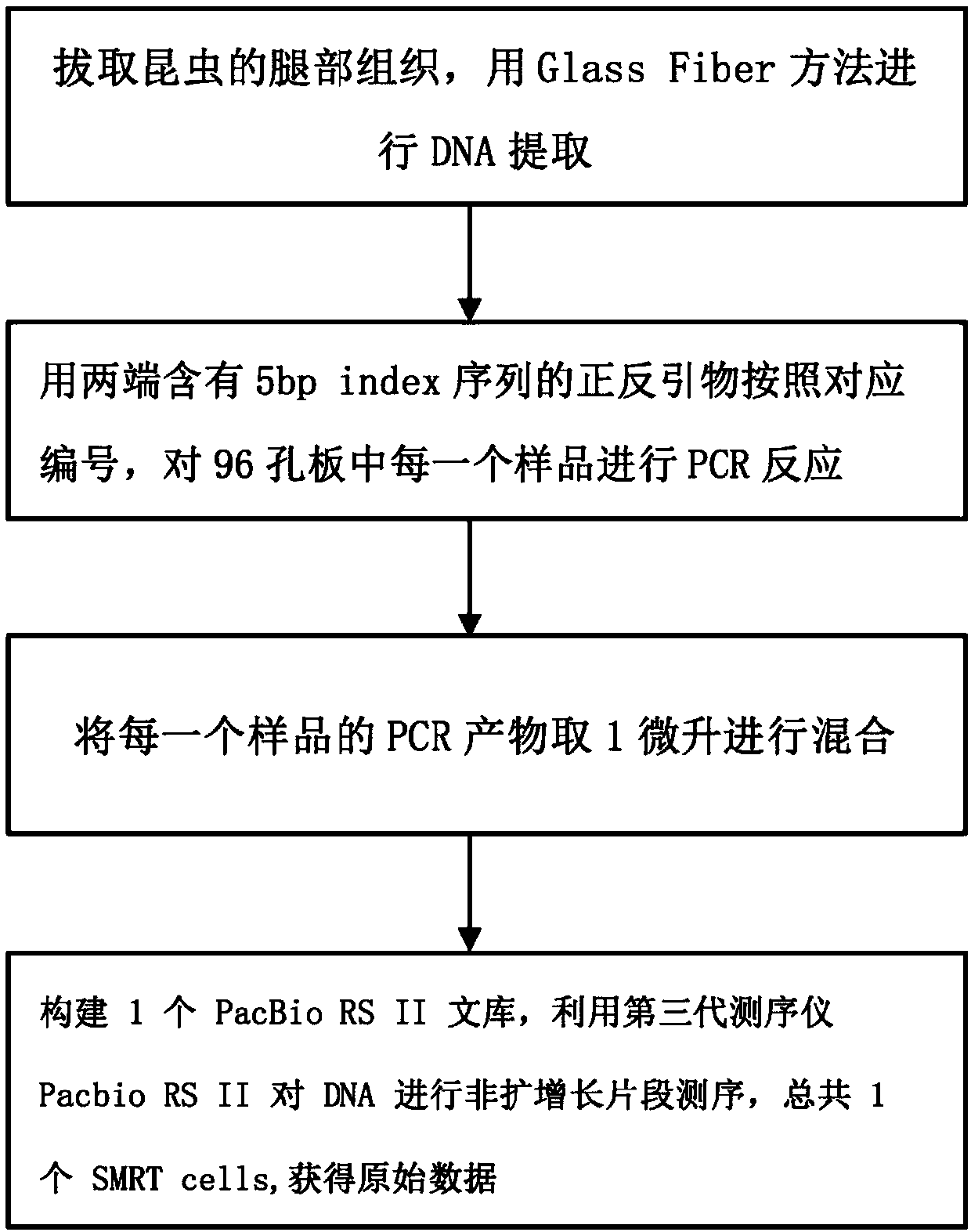
[0061] 1. Extraction of insect DNA
[0062] Insect Lysis Buffer (i.e. "Insect Lysis Buffer+Na 2 SO 3 "), binding mix (ie "BindingBuffer(BB)"), PWB washing liquid and WB washing liquid formulations see:
[0063] http: / / ccdb.ca / site / wp-content / uploads / 2016 / 09 / CCDB_DNA_Extraction-Plants.pdf.
[0064] 1. Clean the test bench with alcohol, and cover it with aluminum foil or plastic wrap to prevent contamination. In order to prevent the static electricity from making the insect legs jump around, about 30 μl of pure alcohol should be added to each well of the 96-well plate first. According to the sample information sheet, add 95 insect samples to 95 wells of a 96-well plate with tweezers (sampling principle: for very tiny insects, the whole body should be taken, multiple legs can be taken for tiny insects, and one leg can be taken for small insects For la...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com