A sybr GreenⅠ fluorescent quantitative PCR kit for detecting Salmonella pullorum and its application
A Salmonella, fluorescent quantitative technology, applied in the field of biotechnology detection, to achieve the effect of simple operation, prevention of mistaken elimination, and high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0037] Example 1: Primer Design
[0038] A comprehensive bioinformatics analysis was performed on the whole genome of pullorum, typhoid and Salmonella enteritidis serotypes in GenBank, and finally the SEEP17695 gene (shown in SEQ ID NO.1) was determined as the detection target gene of pullorum pullorum.
[0039] According to the SEEP17695 gene, specific primers were designed using Oligo 6 software, and the primers were synthesized by Suzhou Jinweizhi Company. The primer sequences are as follows:
[0040] SEEP17695-idF10: 5'TCTAGCACTGAACTTGGCGA 3' (shown in SEQ ID NO.2);
[0041] SEEP17695-idR10: 5'TGTGTCGCCATTGTAGGTCA 3' (shown in SEQ ID NO.3).
Embodiment 2
[0042] Embodiment 2: the establishment of PCR detection method for Salmonella pullorum
[0043] 1. Experimental strains and reagents
[0044] The experimental strains are shown in Table 1, among which 13 isolates were isolated and identified by the Animal Bacteriosis Laboratory of Harbin Veterinary Research Institute.
[0045] Premix Ex Taq DNA polymerase and Marker DL2000 were purchased from TaKaRa Company.
[0046] Table 1 Experimental strains
[0047]
[0048]
[0049] Note: ①: China Veterinary Microbiology Collection Management Center; ②: China Medical Bacteria Collection Management Center; ③: China Industrial Microbiology Culture Collection Management Center; ④: American Type Culture Collection; ⑤: Tiangen Biochemical Technology (Beijing) ) Co., Ltd.; ⑥: preserved in this room.
[0050] 2. Method
[0051] 2.1 Selection of the most suitable enrichment solution and template preparation method
[0052] Five different enrichment solutions were used to amplify Salmo...
Embodiment 3
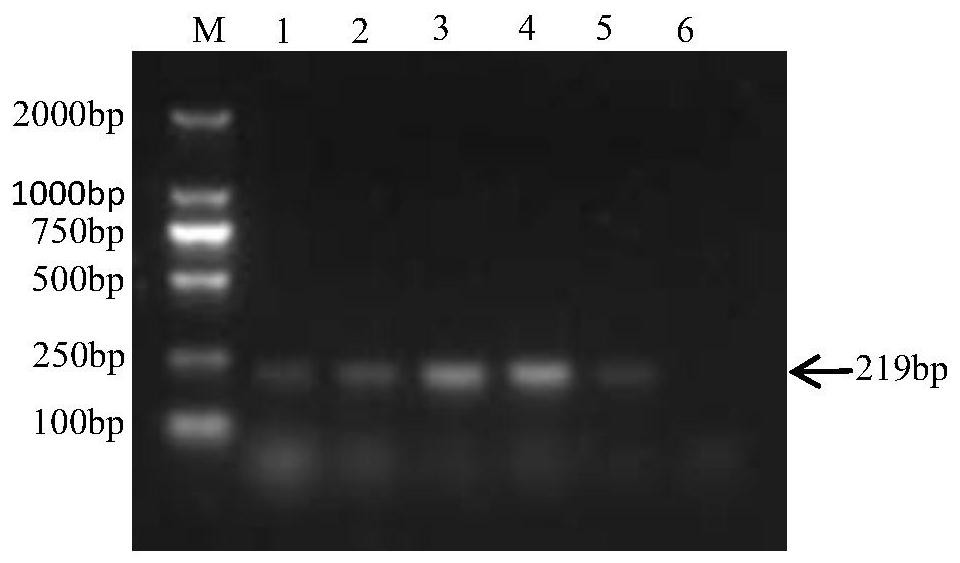
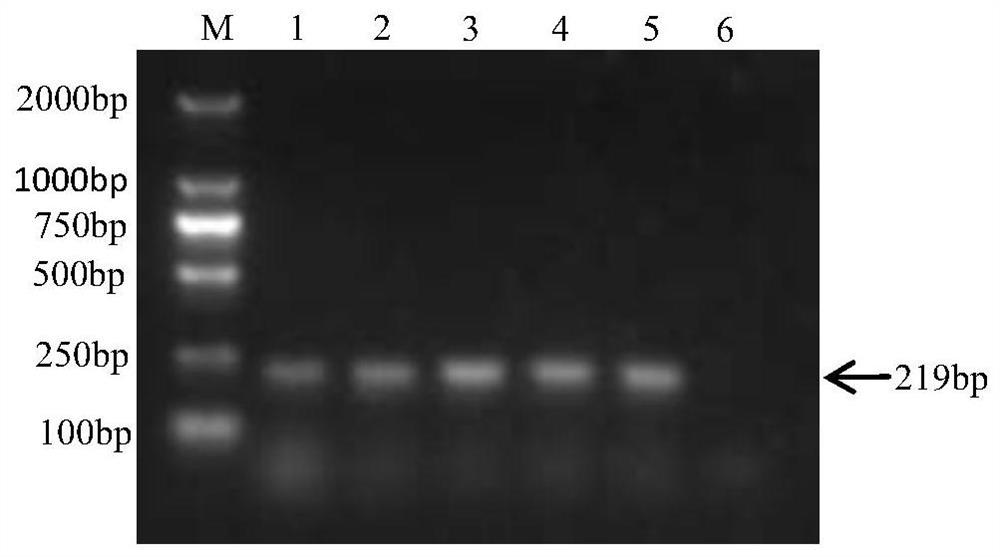
[0078] Embodiment 3: artificial contamination sample detection
[0079] Artificially polluted chicken feces and chicken samples, using the fluorescent quantitative PCR method established in Example 2 to detect Salmonella pullorum, and at the same time using conventional bacterial isolation and identification methods to detect, and evaluate the method established in this study.
[0080] 1. Detection of artificially contaminated chicken feces samples
[0081] Fluorescent quantitative PCR method: Aseptically collect the feces of SPF chickens in the SPF level feeding environment, take 10g of feces and add them to 90mL SC enrichment solution, and cultivate overnight. Dilute the overnight culture of Salmonella pullorum pullorum with sterile water 10 times, and inoculate the appropriate dilution of the bacterial solution into the chicken manure mixture (10g feces + 90mL SC enrichment solution), so that the inoculation amount is 0cfu, 1 ~10cfu, 10~10 2 cfu, 10 2 ~10 3 cfu. Three ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com