Prokaryotic protein acetylation site prediction method
A prediction method and acetylation technology, applied in proteomics, special data processing applications, instruments, etc., can solve the problems of incomplete information on the characteristics of acetylation sites, prediction of acetylation sites of unprokaryotic proteins, etc., and achieve improved prediction Accuracy, effect of dimensionality reduction
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
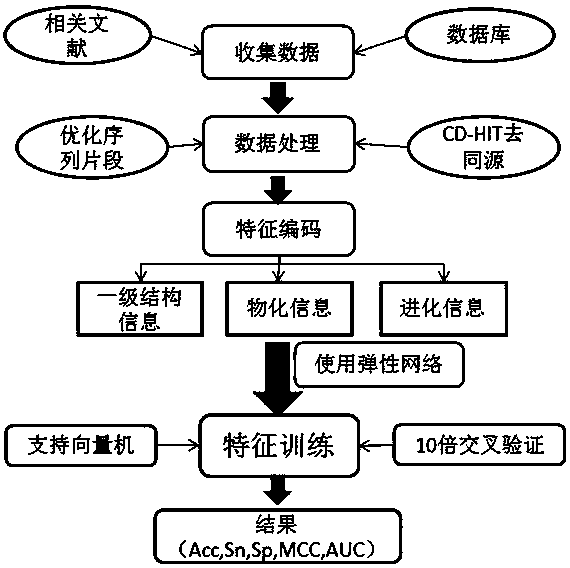
[0064] a kind of like figure 1 The prediction method of the prokaryotic protein acetylation site shown, the specific steps are as follows:
[0065] 1) Collect data
[0066] Collect prokaryotic protein acetylation data from protein databases such as UniProt, CPLM and NCBI and related literature;
[0067] 2) Data processing
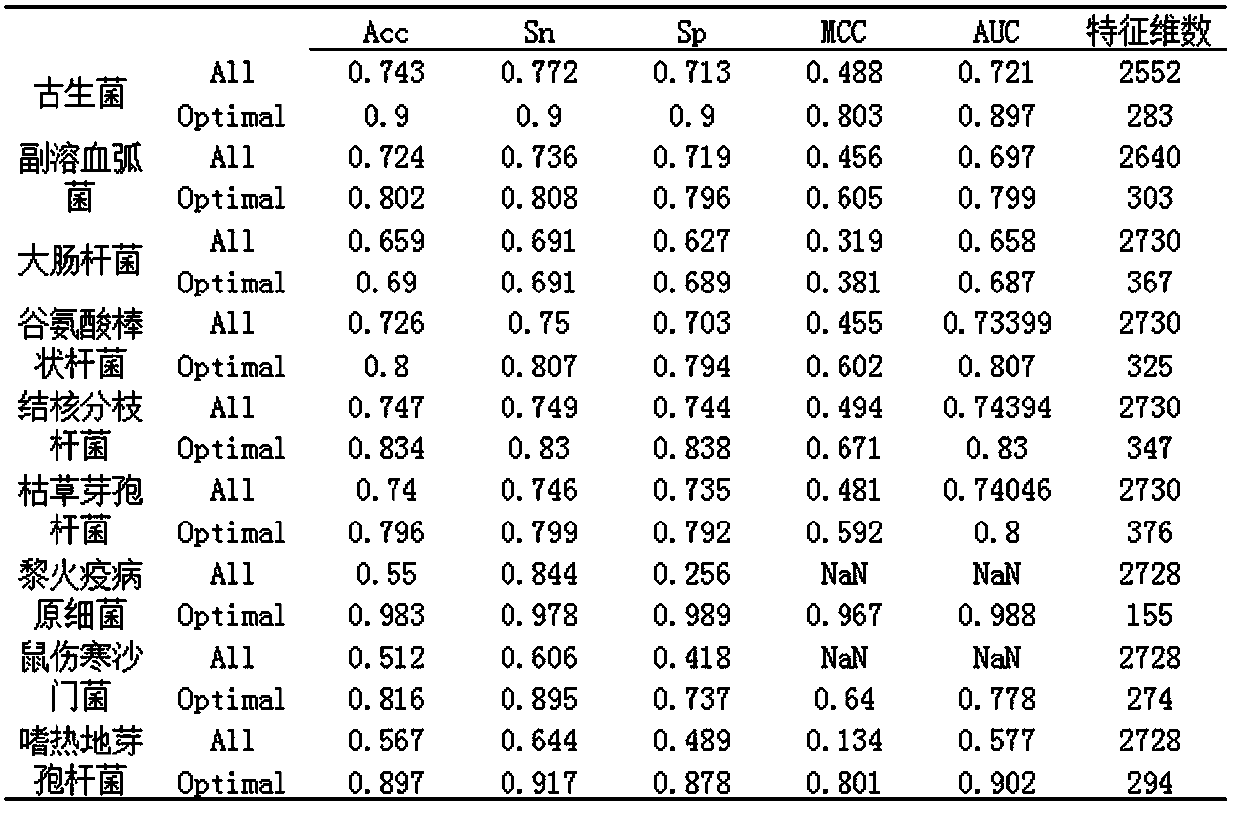
[0068] Archaea, Vibrio parahaemolyticus, Escherichia coli, Corynebacterium glutamicum, Mycobacterium tuberculosis, Bacillus subtilis, Geobacillus thermophile, Lemoniasis et al. A total of nine prokaryotic protein acetylation positive sample data sets and negative sample data sets of Salmonella typhimurium;
[0069] The positive sample is the acetylation site marked by experimental verification, and the negative sample is the unlabeled lysine (K) sequence randomly selected from the same protein as the positive sample with the same number as the positive sample. Data processing includes the following sub-steps:
[0070] 2 ▪1) According to structural bio...
Embodiment 2
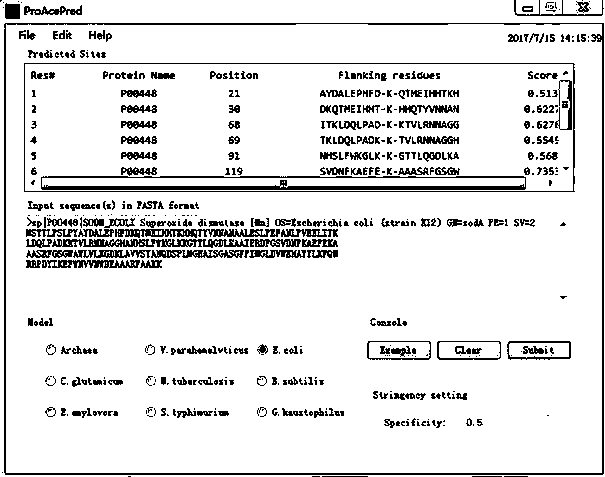
[0105] The prediction software platform ProAcePred was applied to predict the acetylation site of the protein named "P00448".
[0106] The prediction software is the prediction software platform ProAcePred using MATLAB software and C# programming language. The prediction software platform ProAcePred, when the user submits at least one prokaryotic protein sequence, for example, to predict the acetylation site of the protein named "P00448" in the UniProt database, only needs to input the FASTA format of the protein on the prediction interface, select the predicted model and the appropriate Threshold, click the "Submit" button, the ProAcePred tool will predict the "P00448" protein, and automatically give the potential acetylation site information of the protein, and the results will be displayed in the designated area, realizing the acetylation site of prokaryotic proteins High-throughput forecasting.
[0107] image 3 It is the predicted result of lysine acetylation of the seque...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com