Primer composition used for detecting genome target zone and application thereof
A technology of primer composition and target region, applied in the field of primer composition of EGFR mutation site and detection kit, can solve the problems of false positive, unable to completely block the amplification of wild-type DNA, and high cost of reaction reagents, so as to improve the PCR efficiency, solving the effect of low template utilization
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0120] The preparation of embodiment 1 detection kit
[0121] (1) Preparation of primer composition
[0122] Primer probes were designed to detect 23 hotspot mutations of the EGFR gene on the digital PCR platform, and the mutation sites of the EGFR gene are shown in Table 1 below:
[0123] Table 1
[0124]
[0125]
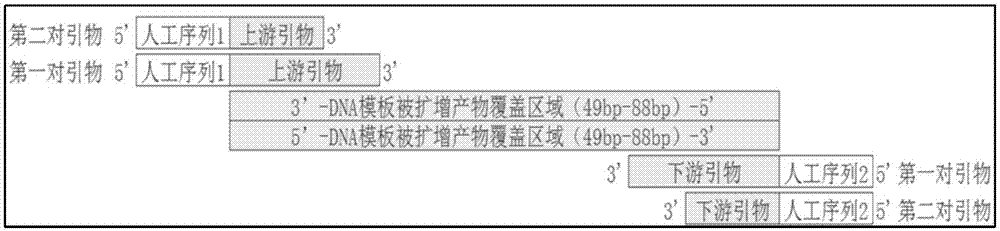
[0126] According to the hot spot mutation of GEFR, design primers such as figure 1 As shown, the specific primer sequences are shown in Table 2 below:
[0127] Table 2
[0128]
[0129] (2) Preparation of detection probes
[0130] According to the hot spot mutation of GEFR, the designed detection probes are shown in Table 3 below:
[0131] table 3
[0132]
[0133] Among them, MT indicates the mutant type, WT indicates the wild type, and bold letters indicate the mutation site;
[0134] (3) Assembly of the kit
[0135] Assemble the primer composition, detection probe and reagents related to the kit to prepare the detection kit.
Embodiment 2
[0136] Example 2 Detection of EGFR site mutation
[0137] The method for detecting EGFR site mutations comprises the steps of:
[0138] (1) Plasma separation: Use an anticoagulant tube to collect 6ml of fresh blood from the patient to separate the plasma within 4 hours, place the blood collection tube at 2000g at 4°C for 10min, take it out gently, transfer the supernatant to a new centrifuge tube, and centrifuge at 16000g at 4°C for 10min , pipette the supernatant into a new 2ml EP tube and store at -80°C;
[0139] (2) DNA extraction: Use 3ml of plasma to extract DNA using Qiagen’s Human Circulating Nucleic Acid Kit (QIAamp Circulating Nucleic Acid Kit) according to the instructions, and dissolve the DNA in 30 μl of AVE buffer or 30 μl of molecular biological water;
[0140] (3) Preparation system:
[0141] (a) The system composition of the EGFR 19del site is shown in Table 4 below:
[0142] Table 4
[0143] components
Final concentration
2×ddPCR Superm...
Embodiment 3
[0162] The EGFR positive standard HD734 purchased from Horizon was mixed with the EGFR negative standard HD709, fragmented by ultrasonic method, used as a simulated sample, and tested at different frequencies and concentrations by gradient dilution. After the DNA template was interrupted, the test results were as follows: Figure 5 shown, from Figure 5 It can be seen that the main peak of the fragment is located between 200bp and 300bp compared with the peak diagram of the known length fragment after the DNA template is interrupted;
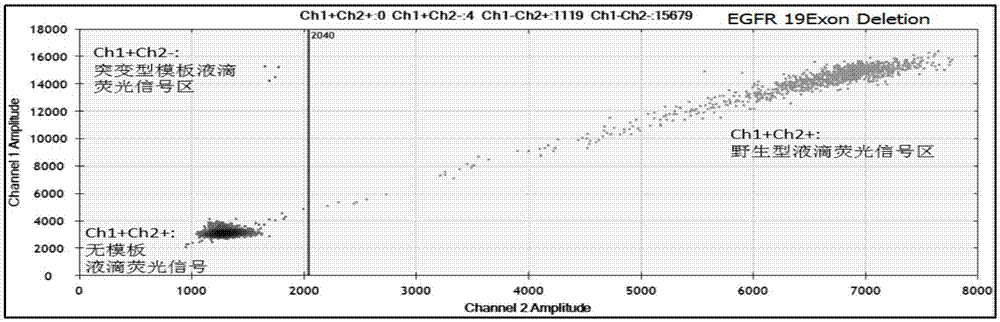
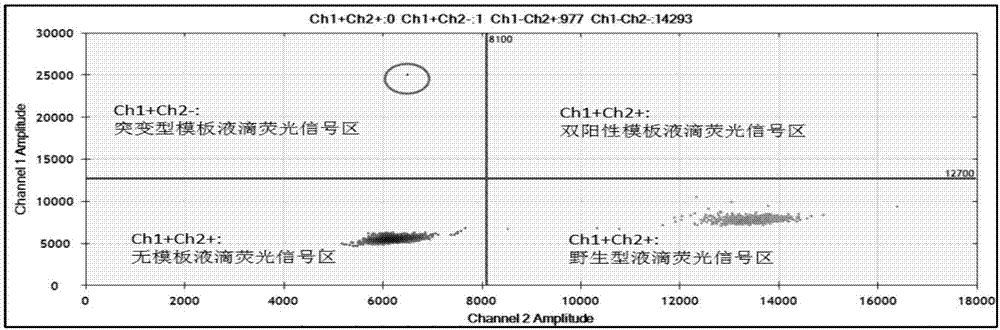
[0163] Using the detection kit in Example 1 to detect with the method in Example 2, the results are shown in Table 9-11, and the two-dimensional diagram of some test results is as follows Figure 6(a)-Figure 6(c) Shown:
[0164] Table 9 EGFR gene exon 19 deletion site
[0165] Amount of DNA (ng)
20
30
50
80
100
120
0.04%
-
+
+
0.02%
+
-
+
0.01%
...
PUM
| Property | Measurement | Unit |
|---|---|---|
| melting point | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com