Method for reducing and/or removing default peptide in solid phase synthesis of polypeptide
A technology of solid-phase synthesis and solid-phase synthesis of peptides, applied in peptide preparation methods, chemical instruments and methods, peptides, etc., to achieve the effects of reducing default peptide impurities, easy access to raw materials, and extensive industrial production value
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0058] The preparation of embodiment 1Fmoc-Gly-Wang Resin
[0059] Weighing 40g of Wang Resin with a degree of substitution of 0.75mmol / g was added to the solid-phase reaction column, washed twice with DMF, swelled with DMF for 30min, washed twice with DMF, and Fmoc-Gly-OH (13.38g, 45mmol) and HOBt ( 6.38g, 47.3mmol) was dissolved in DMF, after ice bath for 10min, DIC (5.95g, 47.3mmol) was added to activate for 3-5min, then added to the reaction column, and DMAP (0.55g, 4.5mmol) was added at the same time, stirred for 2h under nitrogen gas, pumped Dry the reaction solution, wash 4 times with DMF, wash 3 times with DCM, add a DCM solution of acetic anhydride (61.2g, 600mmol) and pyridine (47.5g, 600mmol) to block for 8h, remove the blocking solution, wash 6 times with DMF, and wash 2 times with DCM Once, MeOH was shrunk and dried to obtain 43.6g Fmoc-Gly-Wang Resin with a substitution degree of 0.32mmol / g.
Embodiment 2
[0060] The synthesis of embodiment 2 liraglutide peptide resin
[0061] Weighing 31.3g of Fmoc-Gly-Wang Resin with a degree of substitution of 0.32mmol / g was added to the solid-phase reaction column, washed twice with DMF, swelled with DMF for 30min, washed twice with DMF, and Fmoc-Arg(pbf)-OH( 19.46g, 30mmol) and HOBt (4.26g, 31.5mmol) were dissolved in DMF, ice-bathed for 10min, added DIC (3.97g, 31.5mmol) to activate for 3-5min, then added to the solid-phase reaction column, stirred and reacted for 2h under nitrogen, The ninhydrin detection reaction is complete, the reaction solution is taken out, DMF washes 3 times, 20% DBLK deprotection (5+7min), DMF washes 6 times;
[0062] Dissolve Fmoc-Gly-OH (8.92g, 30mmol) and HOBt (4.26g, 31.5mmol) in DMF, ice-bath for 10min, add DIC (3.97g, 31.5mmol) to activate for 3-5min, then add to the solid-phase reaction column During the reaction, the reaction was stirred with nitrogen for 2 hours, and the reaction was detected by ninhydrin...
Embodiment 3
[0064] Example 3 Liraglutide Peptide Resin Cleavage
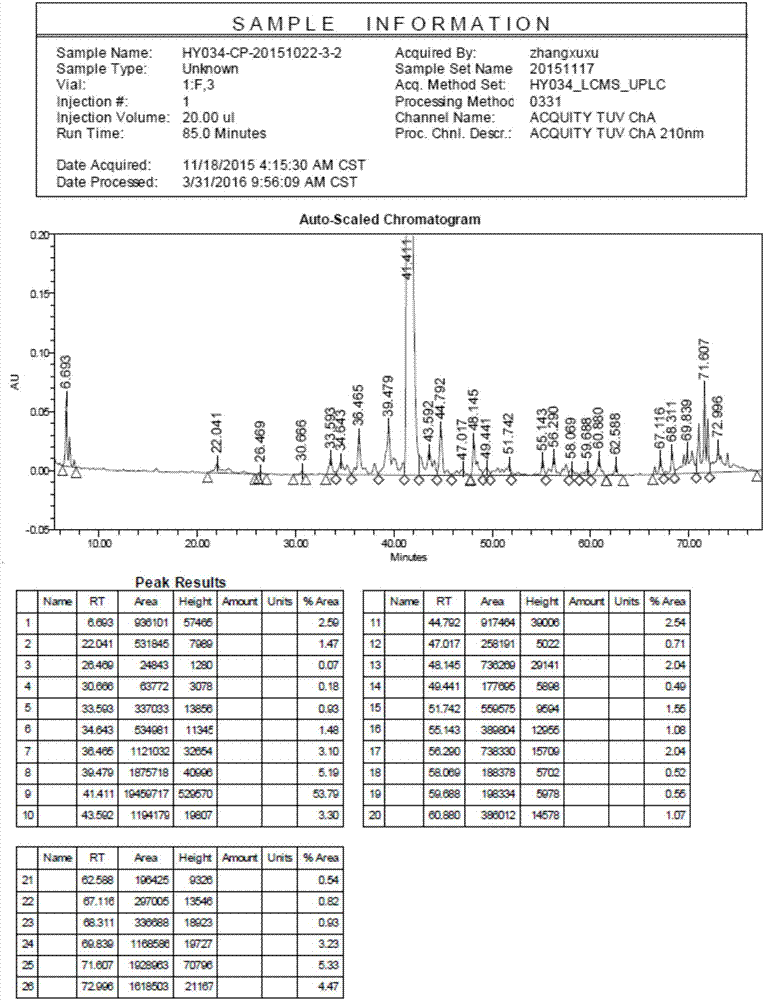
[0065] Transfer 86.2g of liraglutide peptide resin to a 1000ml single-necked round bottom flask, add 860ml of frozen 2h lysate (TFA:Phenol:H 2 O: EDT = 87.5: 5: 5: 2.5), stirred at room temperature for 2 hours, filtered, the filtrate was added to 8.6L of frozen anhydrous ether, centrifuged, washed and dried to obtain 37.3g of crude peptide, yield: 99.5%, purity : 53.79%, see figure 1 .
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com