Method for assisting prediction of risk of occurrence of side effect of irinotecan
一种伊立替康、副作用的技术,应用在辅助对伊立替康副作用的发生风险的预测领域
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0097] [Exploration of related factors of irinotecan side effects]
[0098] Even in patients whose UGT1A gene does not have any side effects-related polymorphisms (UGT1A1*6, *27, *28, UGT1A7 (387T>G, 622T>C), UGT1A9*1b, UGT1A1*60 these 7 places), Serious side effects of irinotecan are often observed. Therefore, the new irinotecan side effects related factors were explored by the following methods.
[0099] (analysis object)
[0100] The next generation sequencer was used to perform exome analysis using genomic DNA prepared from the peripheral blood of the following group: the control group, that is, the 7 polymorphisms mentioned above were all genotypes with low risk of side effects and no side effects were confirmed Case group (group 1: n=5); case group, that is, the 7 polymorphisms of the UGT1A gene mentioned above are all genotypes with low side effects risk, but side effects are still confirmed (Grade): The case group of leukopenia, neutropenia) (group 2: n=5), has a heterozyg...
Embodiment 2
[0110] [Verification of Exome Analysis Results]
[0111] (Verification method of exome analysis results)
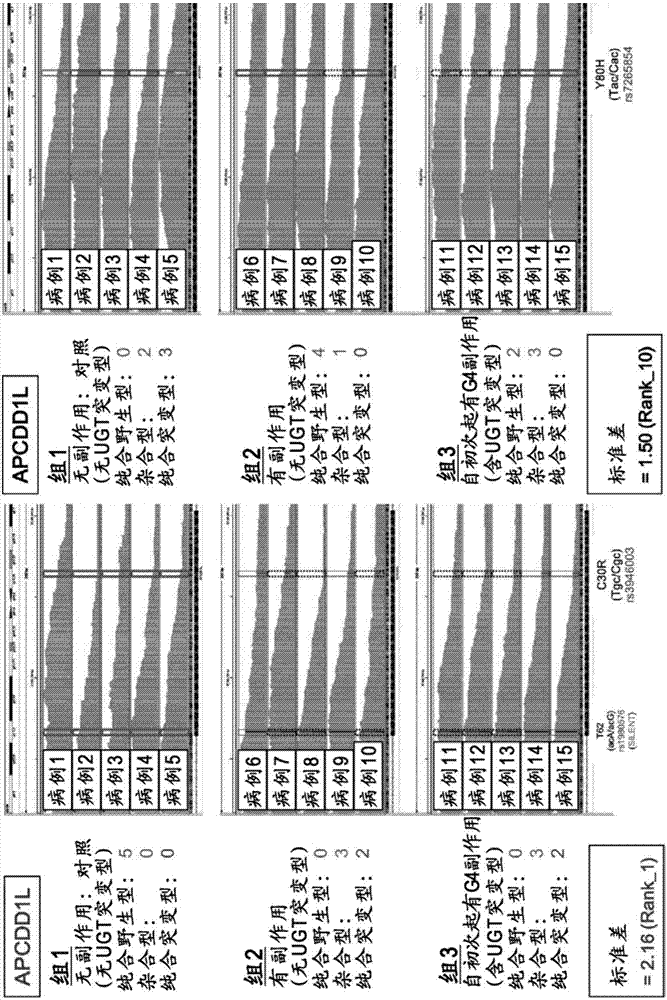
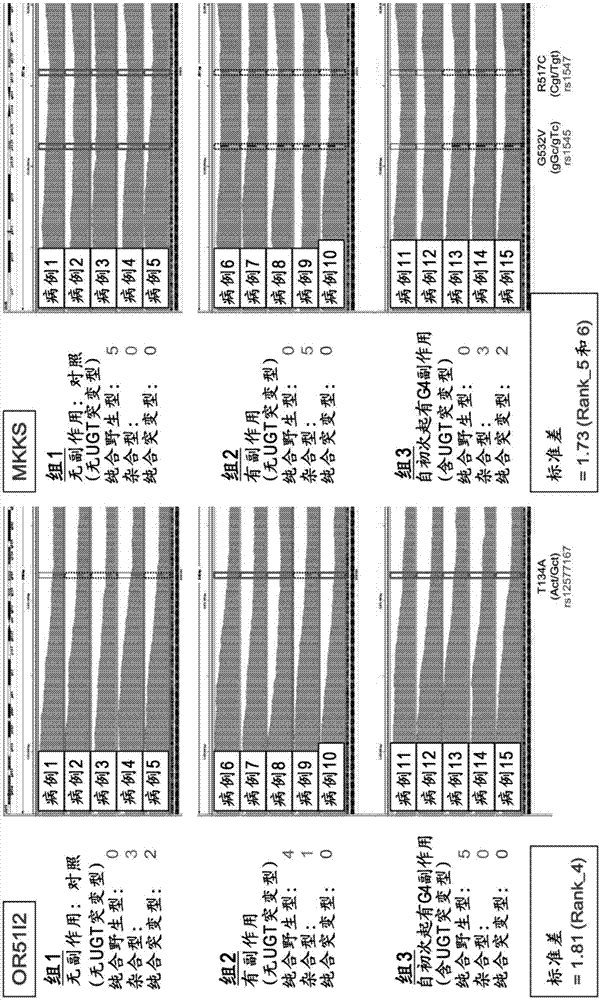
[0112] Among the alternative side effects factors of irinotecan obtained from the results of Example 1, the single nucleotide polymorphism rs1980576 in the region encoding the APCDD1L gene and the single nucleotide polymorphism rs2272761 in the region encoding the R3HCC1 gene The single nucleotide polymorphism rs12577167 in the region encoding the OR51I2 gene, the single nucleotide polymorphism rs1547 in the region encoding the MKKS gene, the single nucleotide polymorphism rs9425343 in the region encoding the EDEM3 gene, and the region encoding the APCDD1L gene The single nucleotide polymorphism rs7265854 in the region encoding the ACOX1 gene, the single nucleotide polymorphism rs1135640 in the region encoding the ACOX1 gene, and the TaqMan (registered trademark) probe method were further used in 75 cases of colorectal cancer patients administered irinotecan (Japan Human) cli...
Embodiment 3
[0116] [Link with UGT1A gene mutation]
[0117] For 74 cases in Example 2, for rs9425343, rs2272761, rs12577167, rs1135640, rs1547, rs7265854, rs1980576, UGT1A9*1b, UGT1A7[387], UGT1A7[622], UGT1A1*60, UGT1A1* as mutations in the UGT1A gene 28. UGT1A1*6 or UGT1A1*27, using TaqMan probe method and direct sequencing method to determine the genotype of each UGT gene, and then using Haploview4.2 software to implement linkage disequilibrium analysis and LD analysis. Show the result in Picture 10 . Picture 10 Where the value is the correlation coefficient (r 2 ). Such as Picture 10 As shown, for rs9425343, rs2272761, rs12577167, rs1135640, rs1547, rs7265854, and rs1980576, no linkage (correlation) with the UGT1A gene mutation was confirmed. In addition, no linkage has been confirmed between rs9425343, rs2272761, rs12577167, rs1135640, rs1547, rs7265854, and rs1980576. It can be seen that these single nucleotide polymorphisms (label sites) can complementarily be used to assist irino...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com