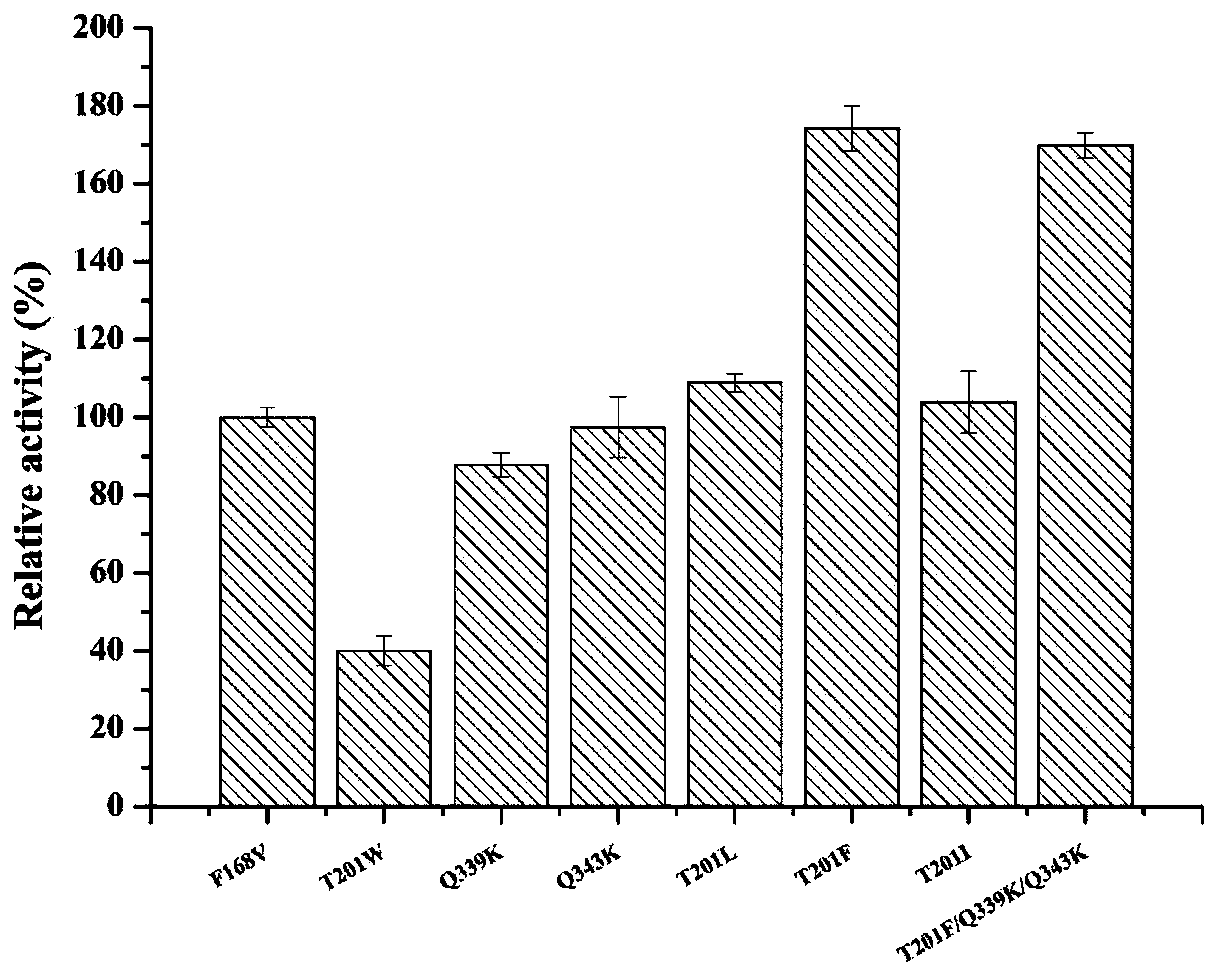
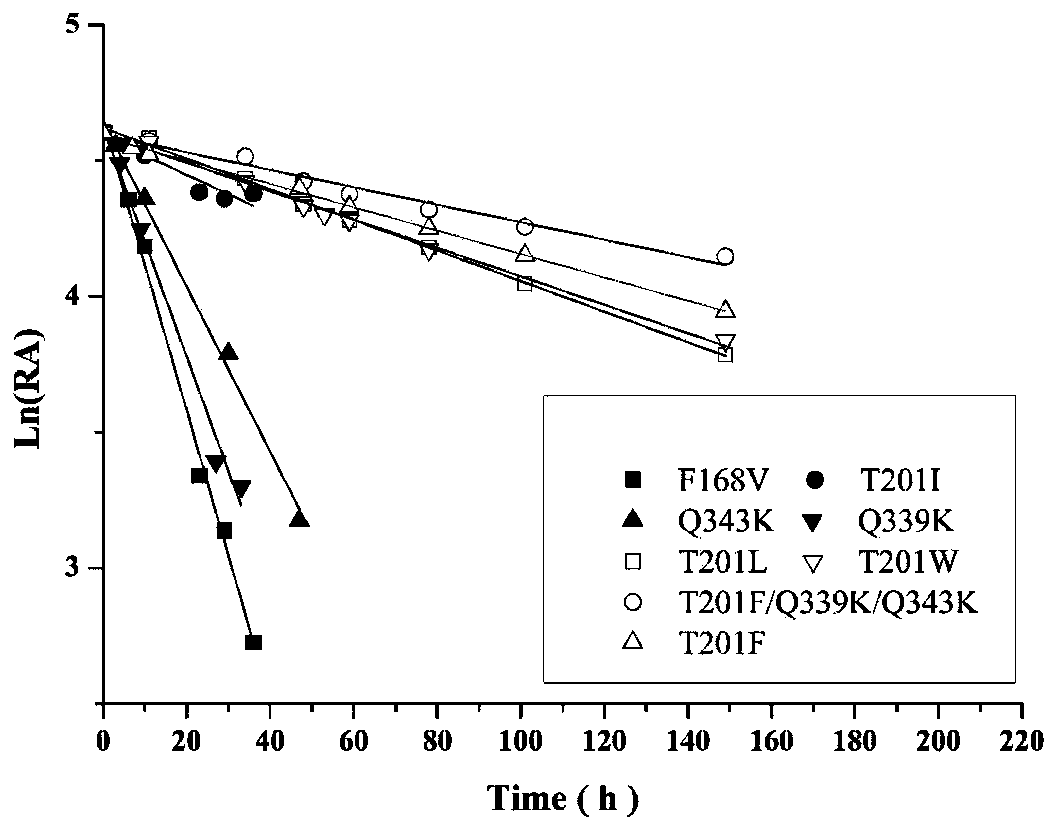
Nitrilase mutant and its application
A technology of nitrilase and mutants, applied in nitrilase mutants and its application fields, can solve the problems of low reaction efficiency, long reaction time, poor temperature stability, etc., achieve high application value, and improve the activity of cyanocarboxylic acid Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] Example 1: Construction of a nitrilase mutant library
[0032] The pET-28b ( +)-AcN-F168V plasmid was used as a template, and T7promoter and T7terminator were used as primers (Table 1) for PCR amplification, and mutations were randomly introduced. PCR reaction system (50μL): template 0.5~20ng, 1×Taq Buffer (without Mg 2+ ), 0.2mM dNTP, 0.3mMnCl 2 , 2mM MgCl 2 , primers T7promoter and T7terminator each 0.2μM, Taq DNA polymerase 5U. PCR conditions (1) pre-denaturation at 95°C for 5 min; (2) denaturation at 94°C for 50 s; (3) annealing at 55°C for 60 s; (4) extension at 72°C for 120 s, a total of 30 cycles of steps (2) to (4); (5) ) and finally extended at 72°C for 5min, and stored at 4°C. The PCR products were analyzed by agarose gel electrophoresis and recovered by cutting the gel.
[0033] Subsequently, the product recovered from the above-mentioned gel was used as a primer to amplify to obtain a complete plasmid. PCR system (50μL) is 5×PrimeSTAR Buffer (Mg 2+ p...
Embodiment 2
[0038] Example 2: High throughput screening of mutants
[0039] Pick a single colony in Example 1 and culture it in a 96-deep well plate, add 1000 μL LB liquid medium (containing a final concentration of 50 μg / mL kanamycin, and a final concentration of 1 mM IPTG) to each well plate, and incubate at 37° C. for 18 hours. Centrifuge the cells in the 96 deep-well plate for 30 min (3000 rpm, 4°C), discard the supernatant, and resuspend the cells with 1.5 mL of sodium dihydrogen phosphate-disodium hydrogen phosphate buffer (200 mM, pH 7.0). Take 500 μL of bacterial suspension into a new 96-deep well plate, add substrate 1-cyanocyclohexylacetonitrile (final concentration 100 mM) to each well, and react at 35° C. for 1 h. Take 500 μL of the bacterial suspension into a new 96-deep well plate, add the substrate 1-cyanocyclohexylacetic acid (final concentration 100 mM) to each well, and react at 35° C. for 12 h. The rest of the bacterial solution was placed in a constant-temperature wat...
Embodiment 3
[0042] Example 3: Site-directed mutagenesis and screening
[0043] Using the expression plasmid pET-28b(+)-AcN-F168V as a template, site-directed mutagenesis was performed by amplification of the whole plasmid. The PCR system (50 μL) is: template 0.5~20ng, primers T201-f and T201-r (see Table 1 for sequence) each 10-15pmol, 5×PrimeSTAR Buffer (Mg 2+ plus), 0.2mM dNTPs, 1.25 U PrimeSTAR HS DNA Polymerase. PCR conditions (1) Pre-denaturation at 98°C for 3min; (2) Denaturation at 98°C for 10s; (3) Annealing at 55°C for 5s; (4) Extension at 72°C for 6.3min, steps (2) to (4) totaled 30 cycles; ( 5) Finally, extend at 72°C for 5 minutes and store at 4°C. The PCR product was verified by agarose gel electrophoresis, digested with DpnI, introduced into E.coli BL21(DE3), and applied to an LB plate containing 50 μg / mL kanamycin to obtain a single clone. In this round of site-directed saturation mutation, a total of 200-300 single clones were generated, which were sequenced and contain...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com