Serum protein marker group for diagnosis of type 1 and type 2 diabetes mellitus
A type 2 diabetes and serum protein technology, applied in the field of diabetes diagnostic protein markers, can solve the problems of diagnosis deviation and poor treatment effect, and achieve reliable and accurate diagnosis results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0035] Differentially expressed proteins in patients with MODY and healthy controls detected by iTRAQ technology
[0036] 1. Test samples:
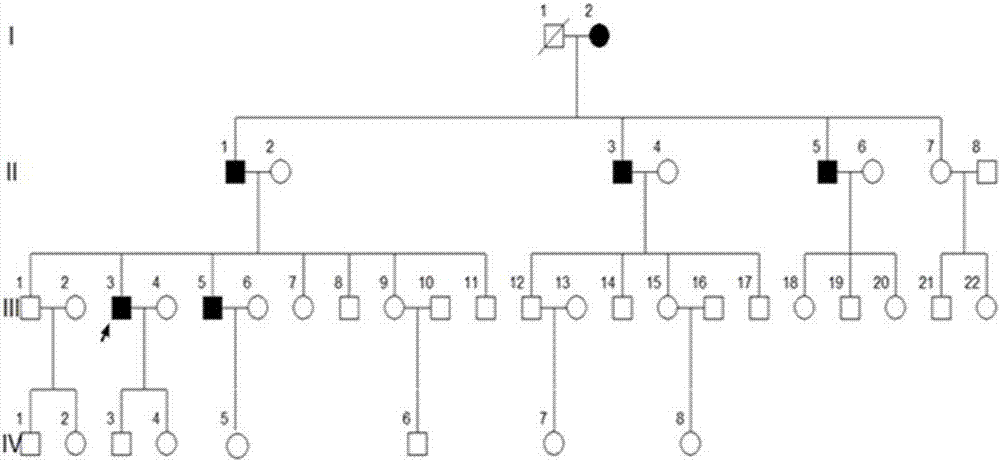
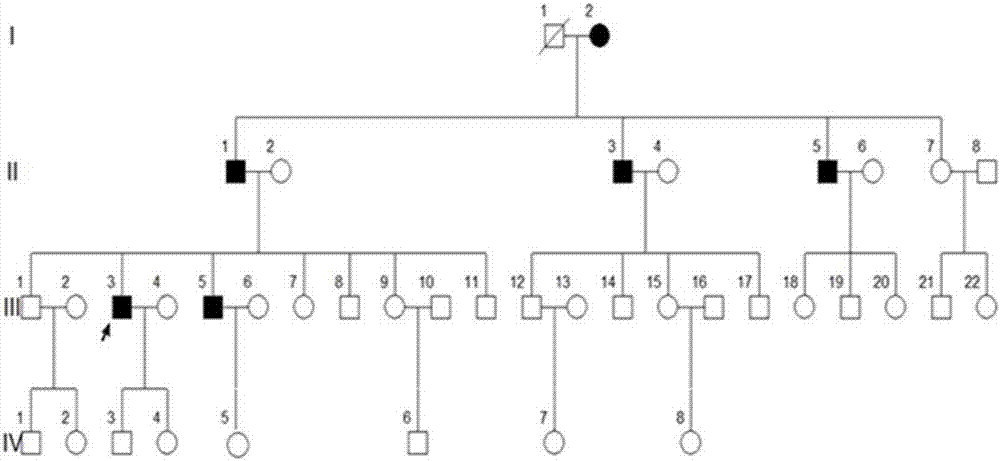
[0037] A four-generation surviving Uyghur family with premature diabetes. Divided into 4 cases of case and 4 cases of normal control serum. Collect 2mL of whole blood on an empty stomach in the morning, let it stand at 4°C for 1-2h until the blood coagulates and precipitate the serum, centrifuge at 3000g for 10min, collect the supernatant, aliquot it on ice and store it at -80°C for later use.
[0038] 2. Detection method:
[0039] (1) Use the Proteominer kit to remove high-abundance proteins in serum: add DTT with a final concentration of 10mM to the enriched protein sample, and bathe in water at 56°C for 1h; after cooling to room temperature, add IAM with a final concentration of 55mM, and place in a dark room for 45min Add 1mL of cold acetone to precipitate overnight, centrifuge at 25000rpm*4°C for 15min and discard the supernatant;...
Embodiment 2
[0059] Validation of Mass Spectrometry MRM in MODY Family
[0060] 1. Test samples:
[0061] MODY patients, 4 cases of normal control serum; all from the family
[0062] 2. Detection method:
[0063] The establishment of the experimental method 1) Select the MODY-related target protein suitable for MRM detection and analysis; 2) Evaluate the quality of the extracted protein; 3) Select the parent-child ion pair suitable for MRM detection; 4) Analyze the mass spectrum based on the analysis software Skyline 5) Perform quality assessment and analysis on the obtained detection data. Sample Analysis:
[0064] (1) Serum proteolysis: 12.5ug of each sample was taken, combined into two tubes of MODY case group and healthy control group, and 01ug / ul of BSA was added to each tube as an internal reference. Use an ultrafiltration tube, take the protein solution, centrifuge for 20min, remove the bottom solution, add 100ul TEAB, centrifuge as above, repeat the step 3 times, replace a new ...
Embodiment 3
[0074] 1. Test samples:
[0075] Type 1 diabetes patients, type 2 diabetes patients, 10 cases of healthy control serum;
[0076] 2. Detection method:
[0077] The establishment of the experimental method 1) MRM detection and analysis of the selected parent-child ion pairs in the expanded 30 samples; 2) Based on the analysis software Skyline, the mass spectrometer scanning parameters-collision energy were optimized; 4) The obtained detection Data quality assessment and analysis.
[0078] Sample Analysis:
[0079] (1) Serum proteolysis: 200ug was taken from each sample, and 0.1ug / ul BSA was added to each tube as an internal reference. Add DTT with a final concentration of 10mM to the taken original serum sample, and put it in a water bath at 56°C for 1h; after cooling to room temperature, add IAM with a final concentration of 55mM, and place it in a dark room for 45min; add TEAB with a final concentration of 100Mm, according to the protein:enzyme=40: Add Trypsin enzyme at a ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com