Method for producing 2-phenylethanol under biological catalysis
A technology of biocatalysis and phenylethyl alcohol, applied in the field of bioengineering and biotechnology, can solve the problems of low yield, unstable properties of intermediates, and increased costs
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0052] Embodiment 1: Construction of E.coli / PdhBb bacterial strain
[0053] 1. PdhBb gene sequence synthesis and expression vector construction
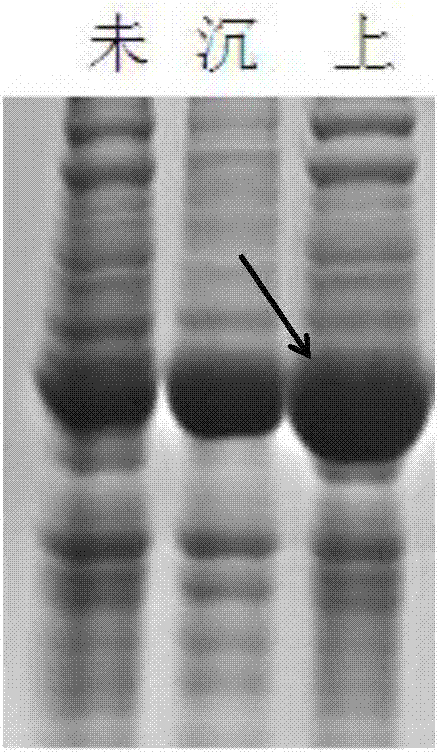
[0054] The PdhBb gene involved in this example has a sequence length of 1143bp, and its accession number in the NCBI database is D45211, which is derived from Bacillus badius IAM 11059, and the sequence is synthesized from the whole gene sequence of Suzhou Jinweizhi Biotechnology Co., Ltd. A nucleic acid restriction endonuclease EcoRI recognition site (5'-GAATTC-3') is added to the 5' end of the fully synthesized PdhBb gene, and a nucleic acid restriction endonuclease NotI recognition site (5'-GCGGCCGC-3') is added to the 3' end ), and cloned into the vector pUC57 to obtain the vector pUC57-PdhBb.
[0055] PdhBb gene target fragment product recovery: Plasmid pUC57-PdhBb was digested with restriction enzymes EcoR1 and Not1 for 2 hours, after 1% agarose gel electrophoresis, the target band was excised, and the DNA was recovered with T...
Embodiment 2
[0062] Embodiment 2: Construction of E.coli / KdcA bacterial strain
[0063] The KdcA gene involved in this example has a sequence length of 1644bp, and its accession number in the NCBI database is AY548760, which is derived from Lactococcus lactis, and the sequence is synthesized from the whole gene sequence of Suzhou Jinweizhi Biotechnology Co., Ltd. A nucleic acid restriction endonuclease EcoRI recognition site (5'-GAATTC-3') is added to the 5' end of the fully synthetic KdcA gene, and a nucleic acid restriction endonuclease NotI recognition site (5'-GCGGCCGC-3') is added to the 3' end ), and cloned into vector pUC57 to obtain vector pUC57-KdcA.
[0064] KdcA gene expression vector construction: KdcA target gene digestion treatment, product recovery, ligation transformation, etc. refer to Example 1.
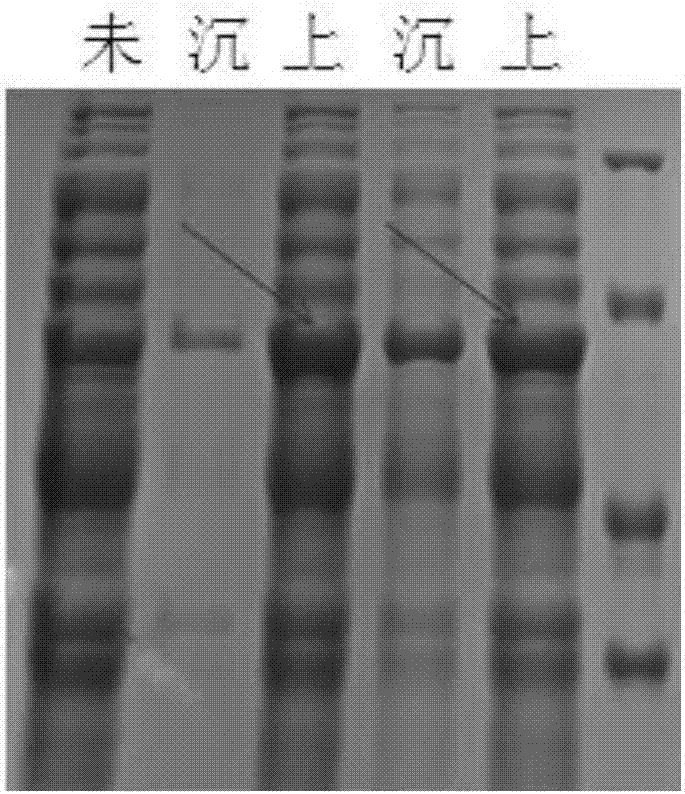
[0065] IPTG induced expression, SDS-PAGE protein expression identification, experimental operation with reference to Example 1, see the results figure 2 . figure 2 The resu...
Embodiment 3
[0066] Embodiment 3: Construction of E.coli / YahK bacterial strain
[0067] The YahK gene involved in this example has a sequence length of 1638bp and its accession number in the NCBI database is 944975, which is derived from Escherichia coli BW25113, and the sequence is synthesized from the whole gene sequence of Suzhou Jinweizhi Biotechnology Co., Ltd. A nucleic acid restriction endonuclease EcoRI recognition site (5'-GAATTC-3') is added to the 5' end of the fully synthetic YahK gene, and a nucleic acid restriction endonuclease NotI recognition site (5'-GCGGCCGC-3') is added to the 3' end ), and cloned into the vector pUC57 to obtain the vector pUC57-YahK.
[0068] Construction of the YahK gene expression vector: refer to Example 1 for enzyme digestion treatment of the YahK target gene, product recovery, ligation transformation, etc.
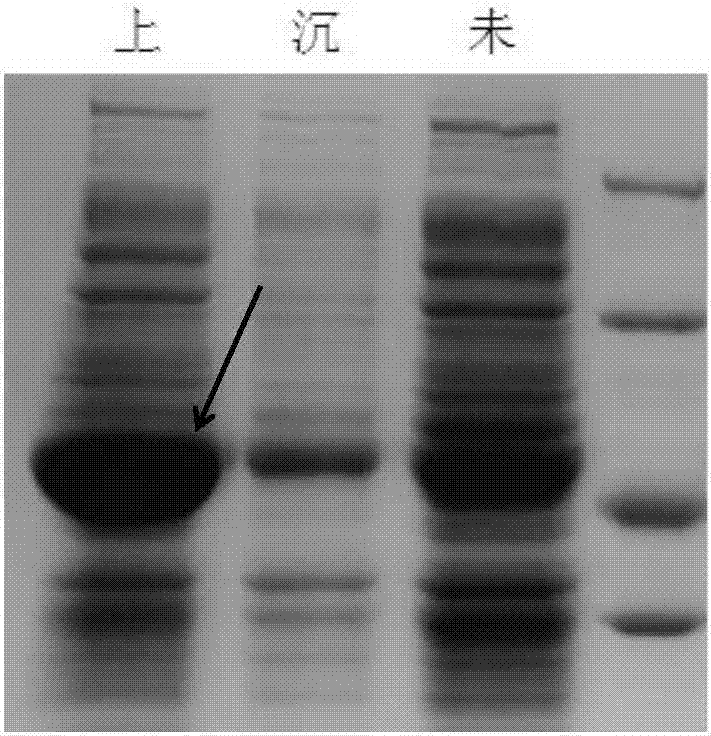
[0069] IPTG induced expression, SDS-PAGE protein expression identification, experimental operation with reference to Example 1, see the resul...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com