Aureobasidium pullulans genome scale metabolism network model and application thereof
A pullulan, genome-scale technology, applied in the field of systems biology
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] Example 1, Construction of Genome-Scale Metabolic Network Model of Aureobasidium pullulans
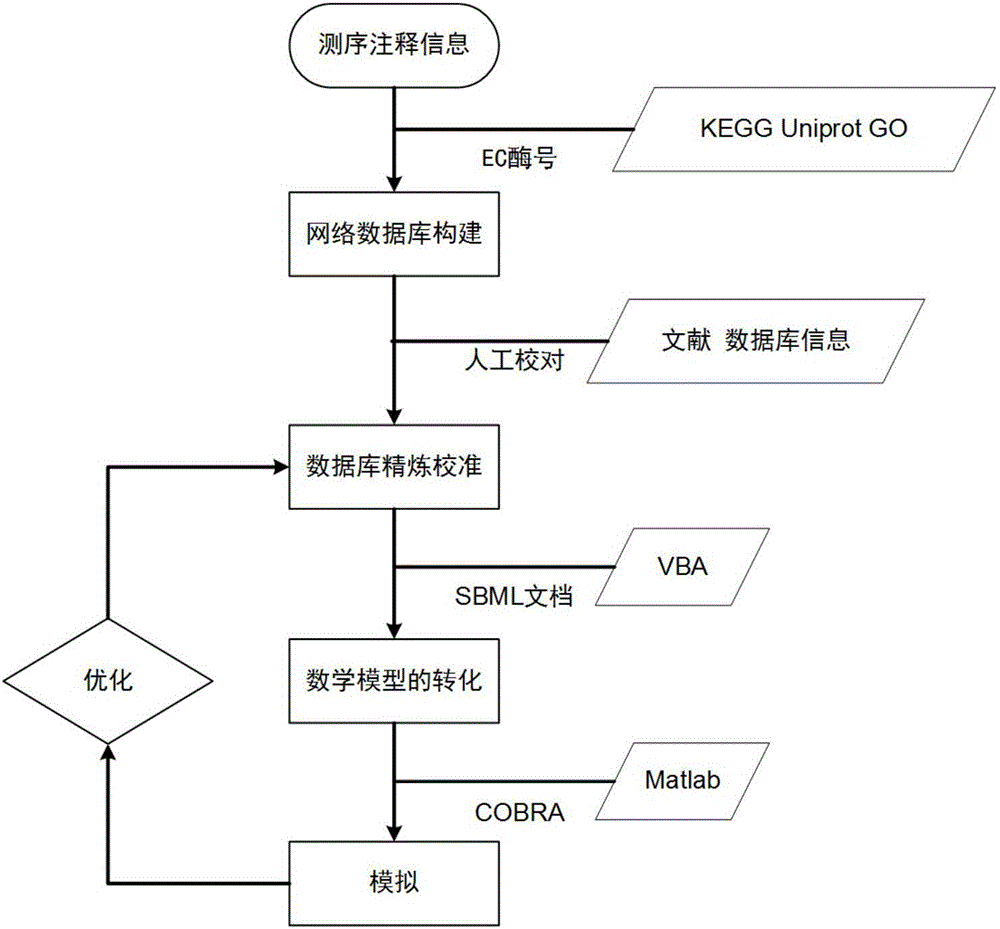
[0033] The construction of the genome-scale metabolic model of Aureobasidium pullulans includes four steps: the establishment of a network database, the refinement and calibration of the database, the establishment of a mathematical model, and the verification and analysis of the model, as follows: figure 1 Shown:
[0034] (1) Establishment of network database: According to the whole genome sequencing results of A. pullulans (A. pullulans) strain CCTCC M2012223 and GO, KEGG, and Uniprot databases, the gene annotation information of A. pullulans was collected, and then the required buds were extracted by VBA program Gene annotation information of Aureobasidium, and then screen out genes, enzymes and reactions related to polymalic acid metabolism, and correlate with each other through EC enzyme numbers to obtain a network database; determine the reaction direction according to the...
Embodiment 2
[0038] Example 2. Features of the genome-scale metabolic network model of Aureobasidium pullulans:
[0039] Based on the above method, the genome-scale metabolic network model iZX637 of Aureobasidium pullulans was constructed. The model contains 637 genes, 1347 reactions and 1133 metabolites, as shown in Table 1, including glycolysis pathway, pentose phosphate pathway, tricarboxylic acid Circulation, pyruvate metabolism, glyoxylate cycle metabolism, amino acid synthesis and consumption metabolism, nucleotide metabolism, lipid metabolism, peptidoglycan synthesis, coenzyme synthesis and other related reactions. Some reactions are listed in Table 2. Allocated into eight cell compartments, all reactions were divided into 9 subsystems according to the KEGG classification standard, and the 9 subsystems were subdivided into 59 metabolic pathways. The model information list is shown in Table 1. Then, based on the strain characteristics and literature information, the synthesis and se...
Embodiment 3
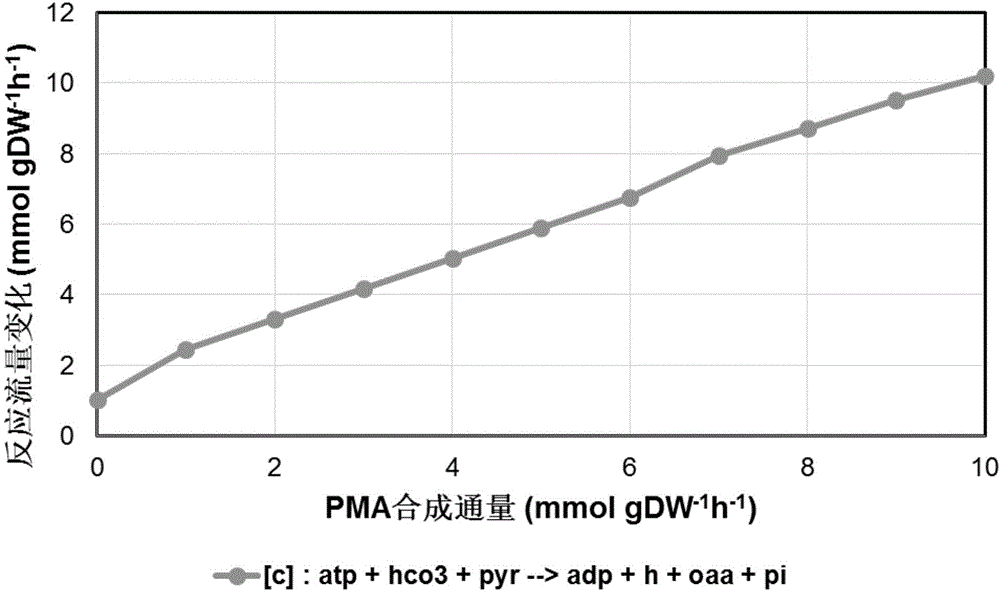
[0048] Example 3. Application of Aureobasidium pullulans genome-scale metabolic network model and target screening
[0049] Using the constructed genome-scale metabolic network model iZX637 of Aureobasidium pullulans, taking the biomass synthesis reaction (reaction ID: biomass) as the objective function, and gradually increasing the polymalic acid secretion reaction (reaction ID: EXC117e), the simulated metabolic flux Unified storage, multiple sets of flow distribution data are obtained, and the reactions with the same trend and the opposite trend with the polymalic acid secretion response are screened out. Then, the biomass and EXC117e reactions were used as the purpose functions to perform single-reaction deletion simulations, and the disturbance results after deletion were counted. The final results showed that under the condition of high PMA synthesis rate, a large amount of carbon flow was concentrated in the pyruvate (PYR) metabolite, which was catalyzed by pyruvate carb...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com