Recombinant corynebacterium glutamicum expressing vgb gene and application of recombinant corynebacterium glutamicum
A Corynebacterium glutamicum, gene technology, applied in the field of genetic engineering, can solve the problems of reduced 4-HIL yield, low yield, complicated process, etc., and achieve the effect of improving 4-HIL yield
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0028] Example 1: Acquisition of vector pJYW-4-ido
[0029] Using the genomic DNA of Bacillus thuringiensis YBT-1520 as a template, primers were designed according to the isoleucine dioxygenase gene ido (GenBank accession no.CP004858.1, locus_tag YBT1520_06070) sequence published by GenBank, and ido-F / ido- R is the primer PCR amplification isoleucine dioxygenase gene ido, and the primers are designed as follows:
[0030] ido-F: 5'-CCGGGGCCCAGAAGGAGGTATAGGATGAAAATGAGTGGCTTTAGCATAGAAGAAAAGG-3'; ido-R: 5'-CGAGGTTAACGTTATTTTGTCTCCTTATAAGAAAATGTT-3'.
[0031] The enzymes used to amplify the target fragments are all high-fidelity PrimerStar DNA polymerases, and the reaction system is 50 μL: 5×PSBuffer 10 μL, dNTP 4 μL, template 1 μL, upstream and downstream primers 0.5 μL, enzyme 0.25 μL, ddH 2 O make up. PCR amplification reaction conditions are: 1. Pre-denaturation at 95°C for 10 minutes; 2. Denaturation at 95°C for 30s; annealing at 55°C for 30s; 1min); the number of cycles is...
Embodiment 2
[0032] Embodiment 2: the acquisition of vector pJYW-4-ido-vgb and recombinant Corynebacterium glutamicum SN01 / pJYW-4-ido-vgb
[0033] Then, using the Genomic DNA of Vitiligo hyaline as a template, design primers according to the sequence of the Vitiligo hyaline hemoglobin gene vgb (GenBank accession no.AF292694.1) published by GenBank, and use vgb-F / vgb-R as primers to amplify the gene vgb by PCR , the primers were designed as follows:
[0034] vgb-F: 5'-ATAGGATCCAGAAGGAGATATACGATGTTAGACCAGCAAACCAT-3';
[0035] vgb-R: 5'-CCTGTCGACTTATTCAACCGCTTG-3'.
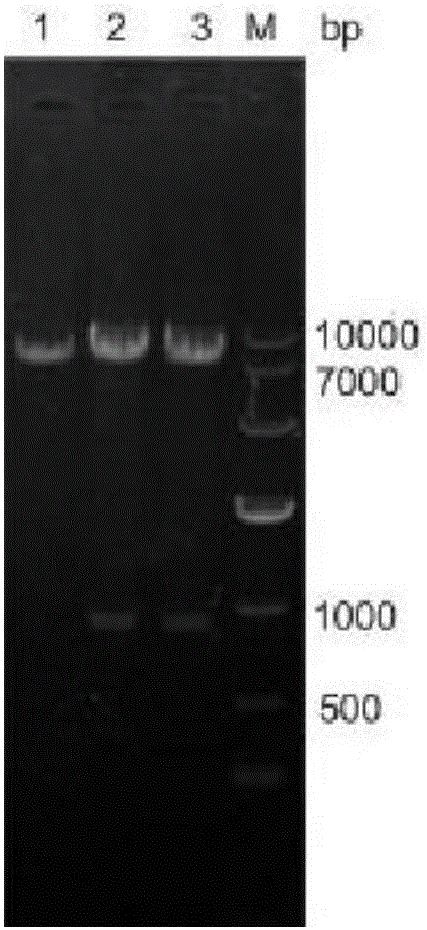
[0036] Using the plasmid pJYW-4-ido as the carrier, repeat the above steps of amplification, digestion, purification, ligation and transformation screening, the fragment vgb and the plasmid were both digested with BamH I and Sal I and purified, then ligated into pJYW- On 4-ido, get pJYW-4-ido-vgb. Figure 4 is the amplified vgb gene map, Figure 5 Shown is the verification map of recombinant plasmid pJYW-4-ido-vgb digestion. ...
Embodiment 3
[0038] Embodiment 3: 4-HIL fermentation of recombinant Corynebacterium glutamicum SN01 / pJYW-4-ido-vgb
[0039] The successful recombinant Corynebacterium glutamicum SN01 / pJYW-4-ido and SN01 / pJYW-4-ido-vgb were compared by shaking flask fermentation.
[0040] The seed medium is: glucose 25g / L, (NH 4 ) 2 SO 4 0.5g / L, urea 1.25g / L, corn steep liquor 40g / L, KH 2 PO 4 1g / L, MgSO 4 0.5g / L, pH 7.0. The seed cultivation temperature was 30° C., and the seeds were cultivated for 18 hours using a rotary shaker at 150 rpm.
[0041] The fermentation medium is: glucose 140g / L, (NH 4 ) 2 SO 4 20g / L, corn syrup 10g / L, KH 2 PO 4 1g / L, MgSO 4 0.5g / L, FeSO 4 0.5g / L, CaCO 3 20g / L, pH 7.2. During the fermentation process, the fermentation culture temperature was 30°C, and the rotary shaker was used to cultivate at 200 rpm for 144 hours. Samples were taken every 12 hours to measure the concentration of bacteria, residual sugar, 4-HIL and other amino acid production.
[0042] Image...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com