Method based on genome LoF loci to screening indirectly related kiwi fruit varietal character
A kiwifruit and genome technology, applied in the field of plant genetic resources evaluation and biology, can solve problems such as difficulty in identifying and utilizing species characteristics, and achieve the effects of improving resource evaluation and utilization efficiency, accurate association prediction, high stability and repeatability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
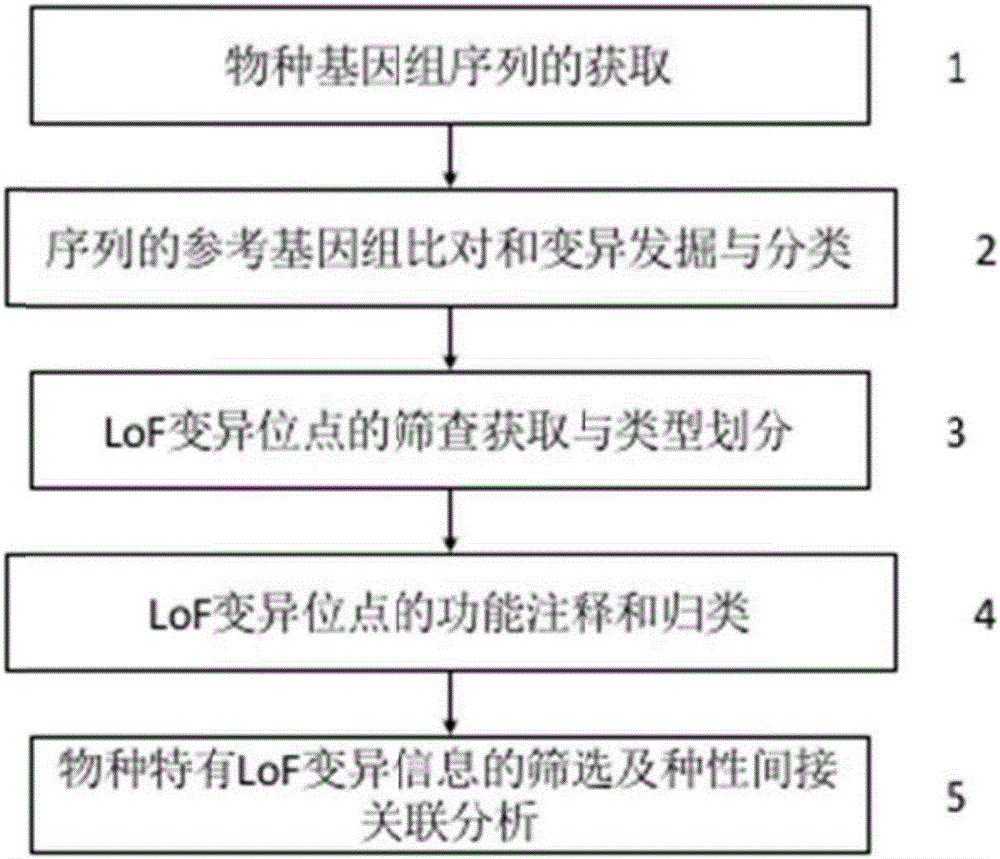
[0024] Embodiment 1: The method for screening indirectly associated kiwifruit species based on the genome LoF site of the present embodiment comprises the following steps:
[0025] 1. If figure 1 Step ① Acquisition of species genome sequence. Using public sequence database resources, including the National Center for Biotechnology Information (NCBI for short), the Kiwifruit Information Resource (KIR for short), and the internal database resources of the National Kiwifruit Germplasm Resource Garden (Wuhan), Obtain short genome sequences of kiwifruit species.
[0026] For kiwifruit species that have not released sequence data, the sequence data (genome short fragment sequence) was obtained by low-depth sequencing of the whole genome. According to the genome size of the Chinese kiwifruit reference genome of about 700M, the second-generation genome sequencing of about 5 times lower depth is performed on the kiwifruit species samples to be sequenced, and the total data volume is ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com