A kit for detecting chicken Newcastle disease virus of different genotypes
A chicken Newcastle disease virus and kit technology, which is applied in the field of general kits for detecting different genotypes of chicken Newcastle disease virus, can solve problems such as high requirements for freshness, and achieve the effects of low cost, good specificity, and overcoming long time-consuming.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0035] Embodiment 1 is used to detect the design of the specific primer of different genotype chicken Newcastle disease virus
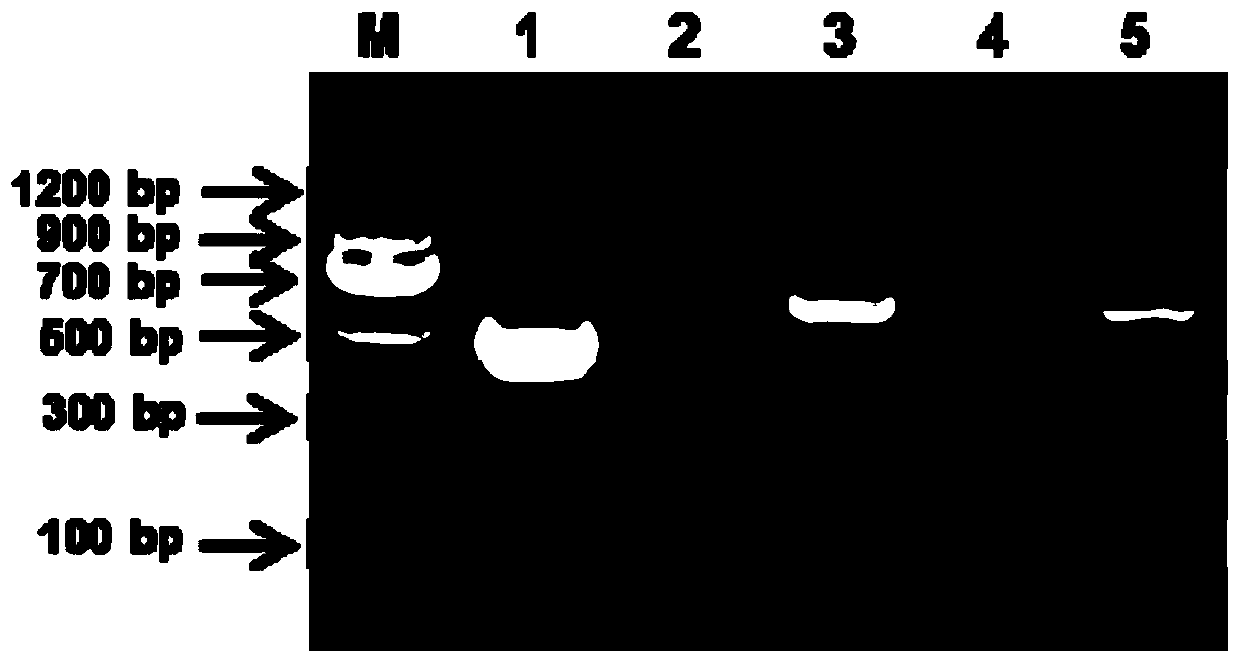
[0036] The genome sequence of NDV-La Sota strain (GenBank accession number: JF950510) in the NCBI database was used as a reference to design specific primers. Among the many alternative primers (shown in Table 1), after repeated screening and comparison tests to exclude possible non-specific matches between the primers and other species sequences, after repeated screening and verification, the optimized primer pair was finally obtained , each primer amplification electrophoresis results are as follows figure 1 shown. Select the primer pair 1 with the brightest amplification target band and no non-specific bands as the best primers. The upstream primer is between 4988nt-5006nt of the accession number JF950510 sequence, and the downstream primer is between 5428nt-5448nt.
[0037] Table 1 Alternative primer sequences for detecting different genotypes o...
Embodiment 2
[0040] Example 2 Detecting Newcastle Disease Virus RT-PCR Detection Method Optimum Annealing Temperature Exploration
[0041] 1. Pretreatment of test samples
[0042] (1) Tissue sample processing: take 100 mg of organ tissue samples, add 0.5 ml of sterilized saline, grind and suspend with a grinder, centrifuge the tissue suspension at 3000 rpm for 30 min, and take the supernatant for detection and analysis.
[0043] (2) Treatment of cloacal or oropharyngeal swab samples: add 0.5 ml sterile saline to the swab sample and oscillate to suspend with a vortex shaker, centrifuge the sample suspension at 3000 rpm for 30 min, and take the supernatant for detection.
[0044] 2. Extraction of total RNA from samples
[0045] Extraction was performed according to the instructions of Trizol RNA Extraction Kit (Invitrogen).
[0046] 3. Reverse transcription into cDNA
[0047] Add the following components into a 0.2ml centrifuge tube: 4 μl of RNA solution, 1 μl of random primers, mix gentl...
Embodiment 3
[0053] Embodiment 3 Detects the RT-PCR detection method of Newcastle disease virus to grope for the optimal number of cycles
[0054] For the pretreatment of the test sample, the extraction of the total RNA of the sample, and the reverse transcription into cDNA, refer to the corresponding method in Example 2. Using the conditions determined in Examples 1 and 2, 6 different cycle numbers (27, 28, 29, 30, 31 and 32) were designed to perform RT-PCR on NDV.
[0055] Add ingredients in 0.2ml centrifuge tube see Example 2. After mixing gently, carry out the following reactions with different cycle numbers: initial denaturation at 94°C for 5min, 94°C for 45s, 54°C for 45s, 72°C for 30s, and different cycle numbers (27, 28, 29, 30, 31 and 32), at the end of the cycle, extend at 72°C for 10 minutes.
[0056] After the PCR reaction, prepare 1% agarose gel with 1×TAE electrophoresis buffer and mix the fluorescent dye Gelsafe according to the reference ratio. Take 7 μl of PCR product a...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com