CRISPR/Cas9 technology-based monocotyledon gene knockout vector and application thereof
A monocot, gene knockout technology, applied in vectors, nucleic acid vectors, genetic engineering, etc., can solve the problems of complex cycle, cumbersome construction process and high cost of plant gene knockout vector system
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
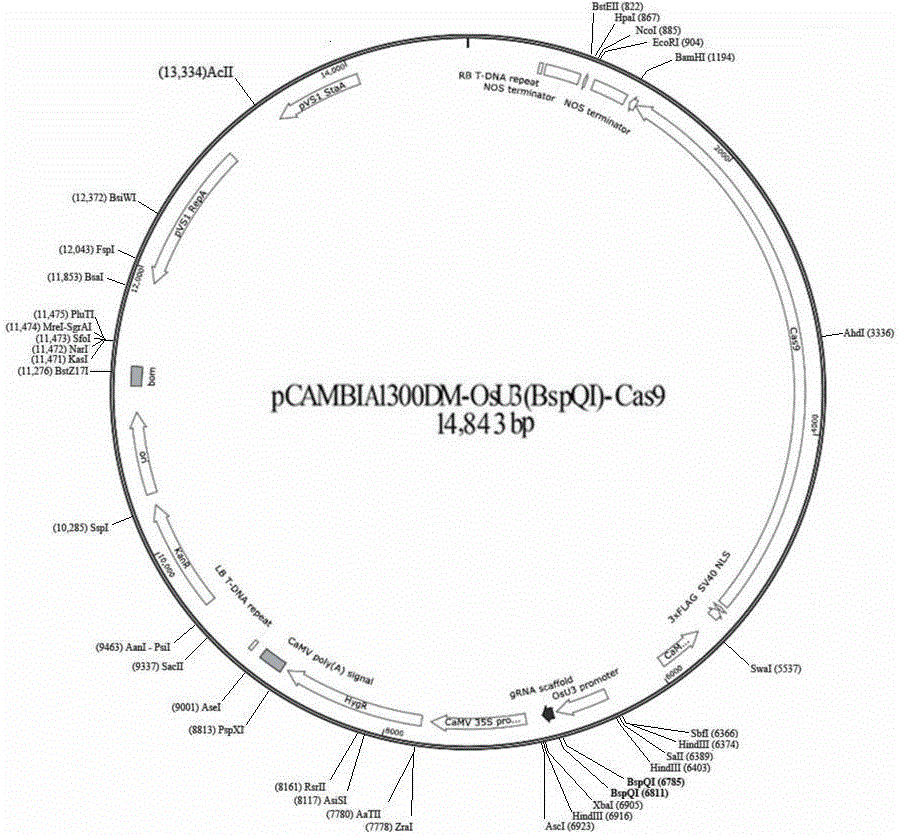
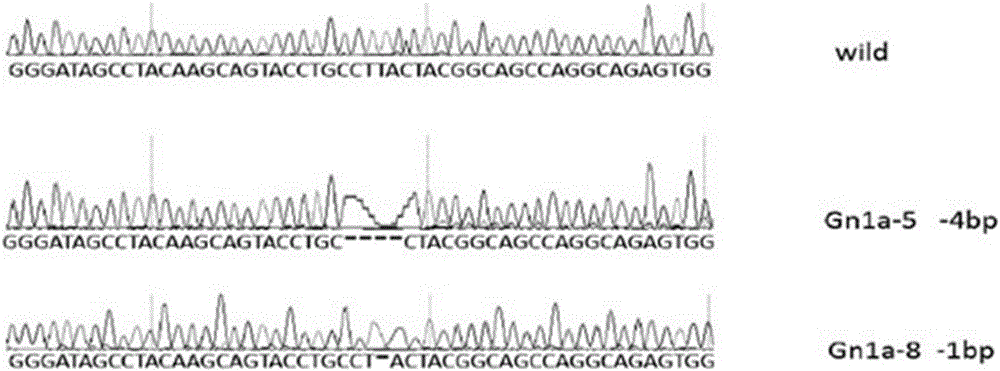
[0047] 1. Construction of rice gene Gn1a knockout vector
[0048] 1. Design a special target sequence according to the rice gene Gn1a sequence and sgRNA design tool, the sequence is as follows:
[0049] Gn1a-oligo1: GGCAGCAGTACCTGCCTTACTA (SEQ ID NO: 10)
[0050] Gn1a-oligo2:AAACTAGTAAGGCAGGTACTGC (SEQ ID NO: 11)
[0051] 2. Perform annealing and phosphorylation of the designed targetoligos according to the following conditions:
[0052] Prepare the reaction system in PCR tubes:
[0053]
[0054]
[0055] Complete the reaction process on the PCR instrument according to the following procedures:
[0056] ①37℃30min
[0057] ②95℃ for 5 minutes
[0058] ③60 cycles:
[0059] 95℃—1.3℃ / cycle 45s
[0060] ④ Store at 4°C
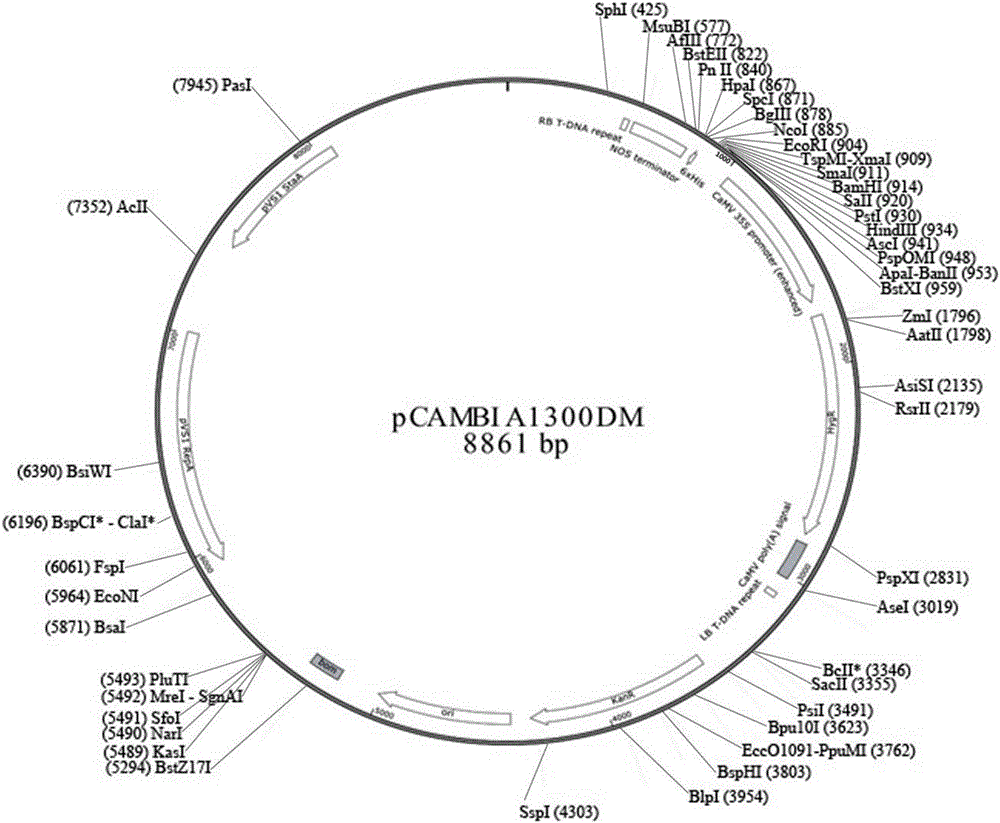
[0061] 3. Digest the pCAMBIA1300DM-OsU3-Cas9 plasmid with BspQ I enzyme to obtain a linearized vector, and use CIP enzyme to phosphorylate it to avoid self-ligation.
[0062] 4. Dilute the annealed phosphorylated product 100 times, and take 2 μl for T4...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com