A method for knocking out the chromosome of Saccharomyces cerevisiae
A technology of Saccharomyces cerevisiae and Saccharomyces cerevisiae strains, applied to methods based on microorganisms, other methods of inserting foreign genetic materials, and stably introducing foreign DNA into the middle direction of chromosomes, which can solve the problem of large workload of complete chromosomes and failure to ensure the integrity of a single chromosome , low efficiency and other issues, to achieve the effect of avoiding cross exchange, high success rate, and rapid knockout
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
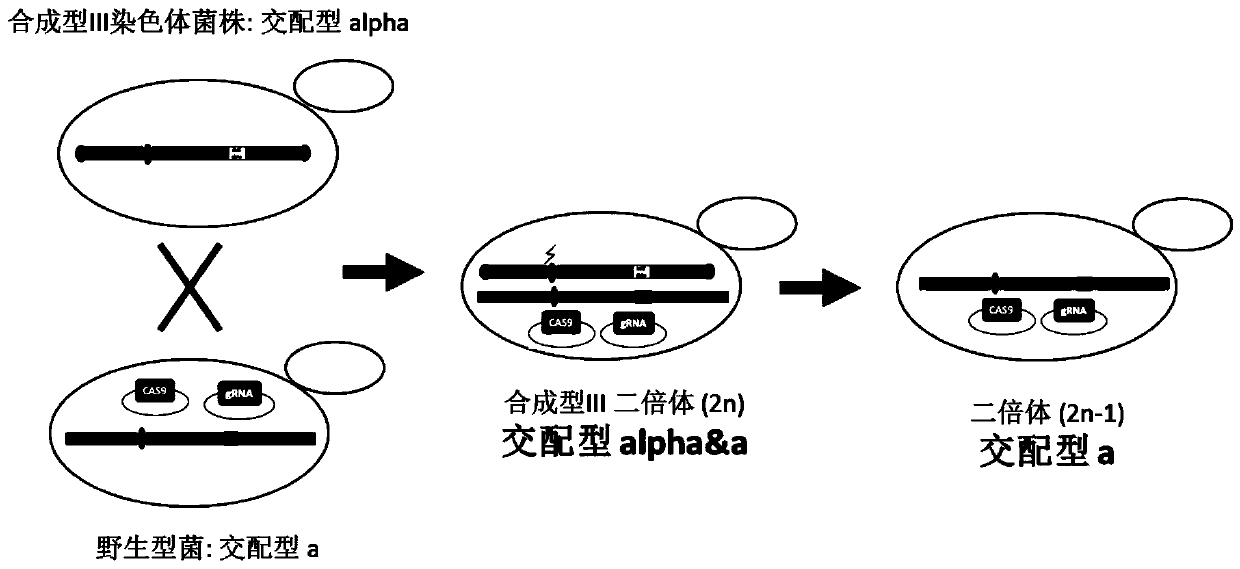
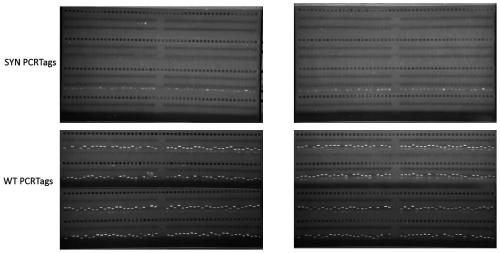
[0029] Example 1: Knockout of synIII artificial Saccharomyces cerevisiae synthetic type III chromosome
[0030]1. The synIII artificial Saccharomyces cerevisiae yYW0233 (MATα, BY4742 based on the artificial synthesis of chromosome III) strain to be knocked out to synthesize chromosome III (synIII) and the yYW0171 (MATa, namely BY4741) strain containing wild chromosome III (wtIII) were brewed Yeast) strains were compared, and the guide RNA recognizable site gRNA-synIII ce n-L "ttatacgaagttattataagCGG" was found near the synI II centromere within 100bp, where CGG is the PAM sequence, and ttatacgaagttattataag is the protospacers sequence.
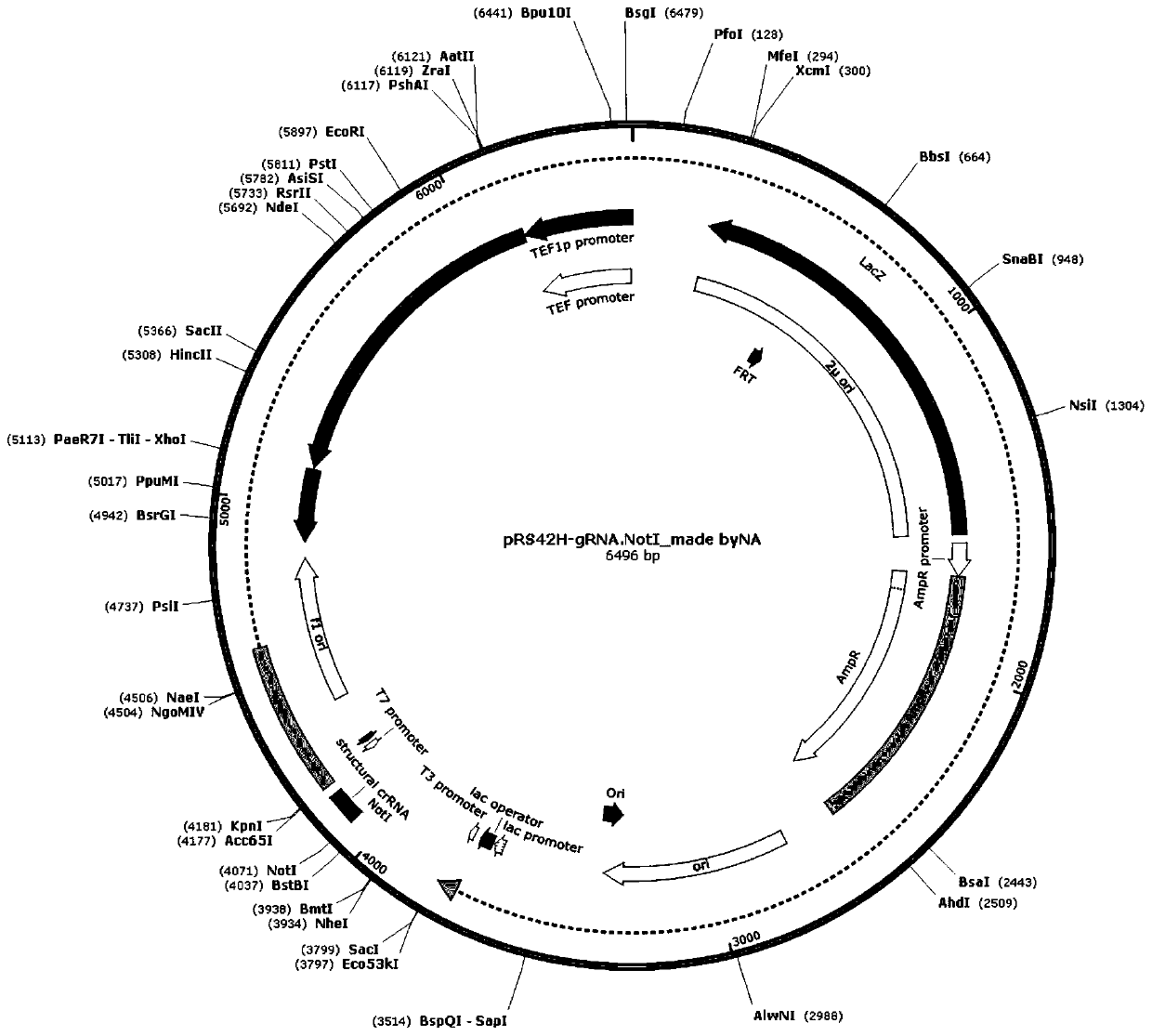
[0031] 2. Construct guide-RNA plasmid, the construction steps are as follows:
[0032] Select the protospacers as ttatacgaagttattataag, and use the NotI restriction site of the pRS42H plasmid as the insertion point;
[0033] Artificially synthesized primers "GCAGTGAAAGATAAATGATCttatacgaagttattataagGTTTTAGAGCTAGAAATAGC" and "GCTATTTCTAGCTCTA...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com