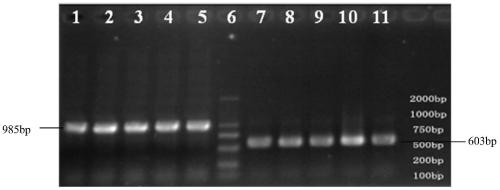
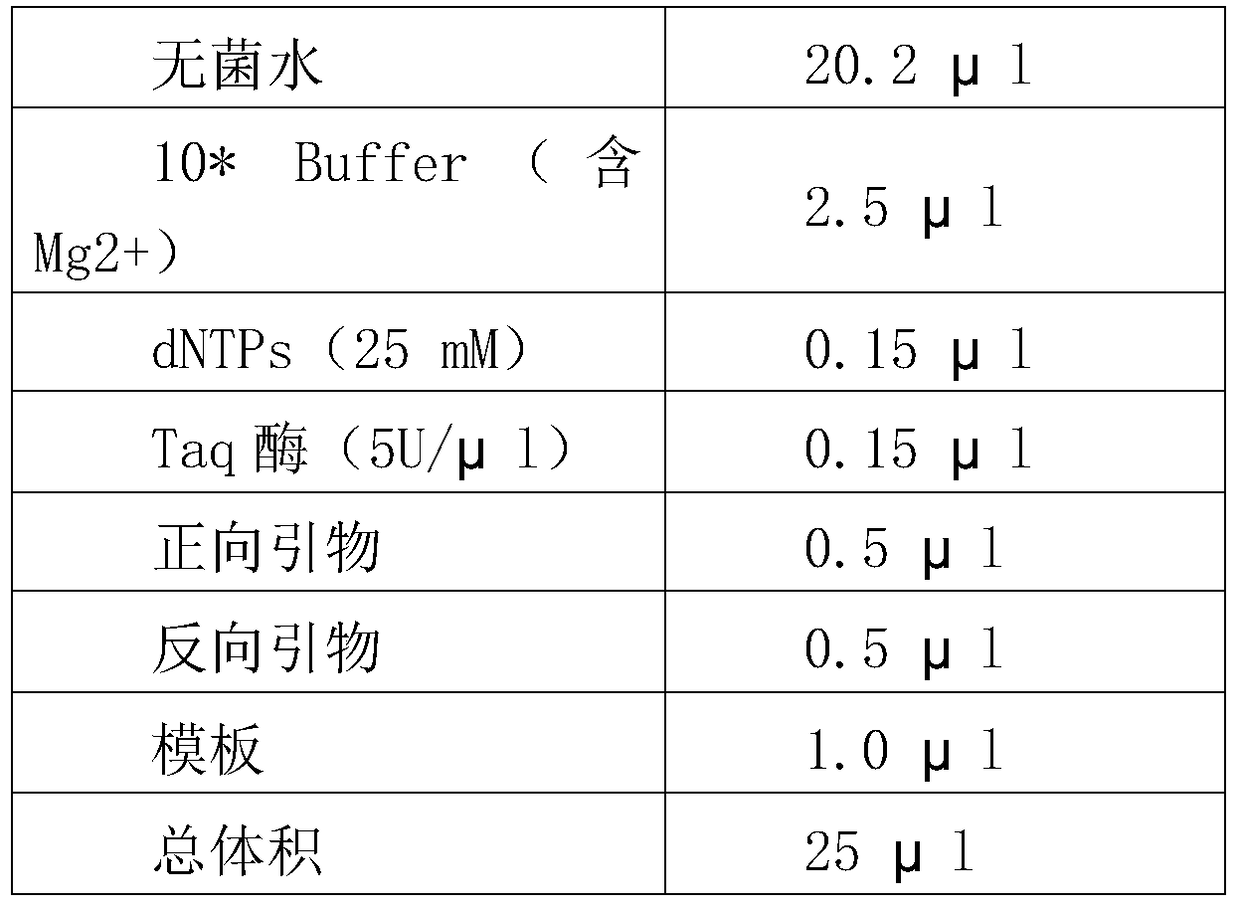
Molecular marker sv3 closely linked to rice bacterial blight resistance gene
A technology of resistance to bacterial blight and molecular markers, applied in the field of molecular biology, can solve the problems of lack of QTLs and molecular markers, complex genetic mechanisms, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0038] Embodiment 1: Construction of rice RIL population
[0039] Rice seeds were purchased from: Shenzhen Huada Agriculture and Circular Economy Technology Co., Ltd.
[0040] Male parent: 801, resistant to bacterial blight;
[0041] Female parent: R15, not resistant to bacterial blight;
[0042] The F1 generation is obtained by crossing the male parent and the female parent;
[0043] 188 recombinant inbred lines RIL populations (F7 generation) were obtained from the F1 generation by the method of single-grain subculture.
Embodiment 2
[0044] Embodiment 2: rice bacterial blight inoculation identification
[0045] A representative strain of dominant pathogenic type IV of bacterial blight of rice in Hunan Province (gifted by Hunan Hybrid Rice Research Center) was selected. The bacterial strain was cultured with Wakimoto's potato semi-synthetic medium at a constant temperature of 28°C for 72 hours, the bacterial lawn was washed with sterile water, and the bacterial suspension was diluted to 108-109 cells / ml bacterial solution by McNairney turbidimetry. Medium formula: 300 grams of potato, 15 grams of sucrose, 5 grams of peptone, Ca(NO 3 ) 2 .4H 2 O 0.5 g, Na 2 HPO 4 .12H 2 O 2.0 grams, agar powder 16 grams, distilled water adjusted to 1000 ml.
[0046] For the RIL population obtained in Example 1, the artificial leaf-cut inoculation method was used to inoculate at the seedling stage of the plant, and 14 to 20 days after the inoculation, when the condition of the susceptible variety was stabilized, the inv...
Embodiment 3
[0050] Example 3: Extraction of Genomic DNA
[0051] Genomic DNA of individual plants of the male parent, female parent, F1 generation and RIL population were extracted by the CTAB method, and the specific methods were as follows:
[0052] (1) Weigh 1.0g of fresh leaves, cut them into pieces and put them in a mortar, grind them with liquid nitrogen, add 3mL 1.5×CTAB, grind them into a homogenate and transfer them to a 15mL centrifuge tube, then add 1mL 1.5×CTAB into the mortar Rinse and transfer to a centrifuge tube. After mixing, place in a water bath at 65°C for 30 minutes, and shake slowly from time to time during this period.
[0053] Among them, the formula of 1.5×CTAB is as follows (1L):
[0054] CTAB
15g
1mol / L Tris.Cl (pH is
75mL
[0055] 8.0)
0.5mol / L of EDTA
30mL
NaCl
61.4g
[0056] Add deionized water to make up to 1 L, and add mercaptoethanol with a final concentration of 0.2% (2 ml) before use. ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| length | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com