A double-colour fluorescence detecting method, primers and probes for rapidly distinguishing Genotype II PRV, Genotype I PRV and a vaccine strain
A two-color fluorescence and fluorescence detection technology, which is applied in the determination/inspection of microorganisms, biochemical equipment and methods, DNA/RNA fragments, etc., can solve the problems of inability to identify strain types, lagging antibody detection, unfavorable vaccine immunity, etc. Achieve fast detection speed, high throughput and good repeatability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0047] Example 1 Primers and Probes
[0048] After screening a large number of designed primers and probes, it was found that the primer pair F, R, and probes P1 and P2 had the best effect on distinguishing porcine pseudorabies virus Chinese type, European and American type and vaccine strain Bartha-K61 by two-color fluorescence method. Well, its base sequence is as follows.
[0049] Primer F: 5'-GGGCGTCTACACGTGGCGC-3' (SEQ ID NO: 1),
[0050] Primer R: 5'-GTTGGTCACGAAGGCGGCGT-3' (SEQ ID NO: 2),
[0051] Probe P1: 5'-FAM-CGCAGCGCCAACGTCTCGCTCGTCCTGTAC-BHQ1-3' (SEQ ID NO: 3),
[0052] Probe P2: 5'-HEX-CCGAGTTCGGCCTGAGCGCGCCGCC-BHQ1-3' (SEQ ID NO: 4).
Embodiment 2
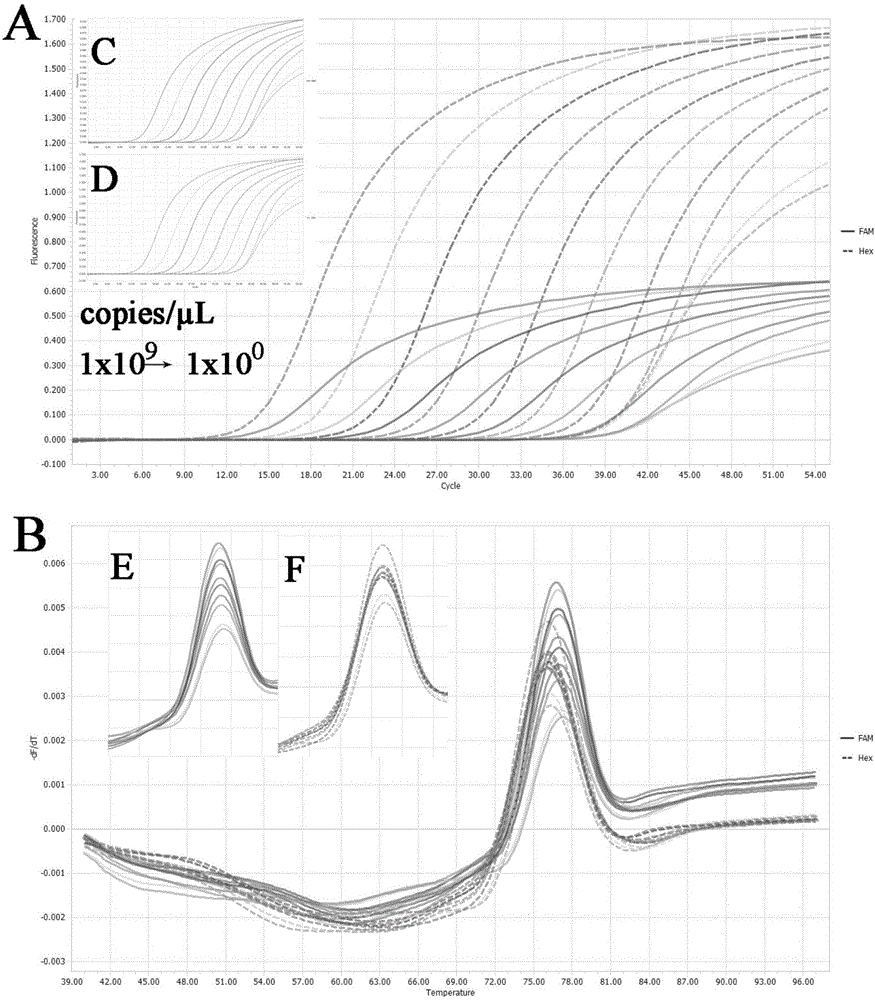
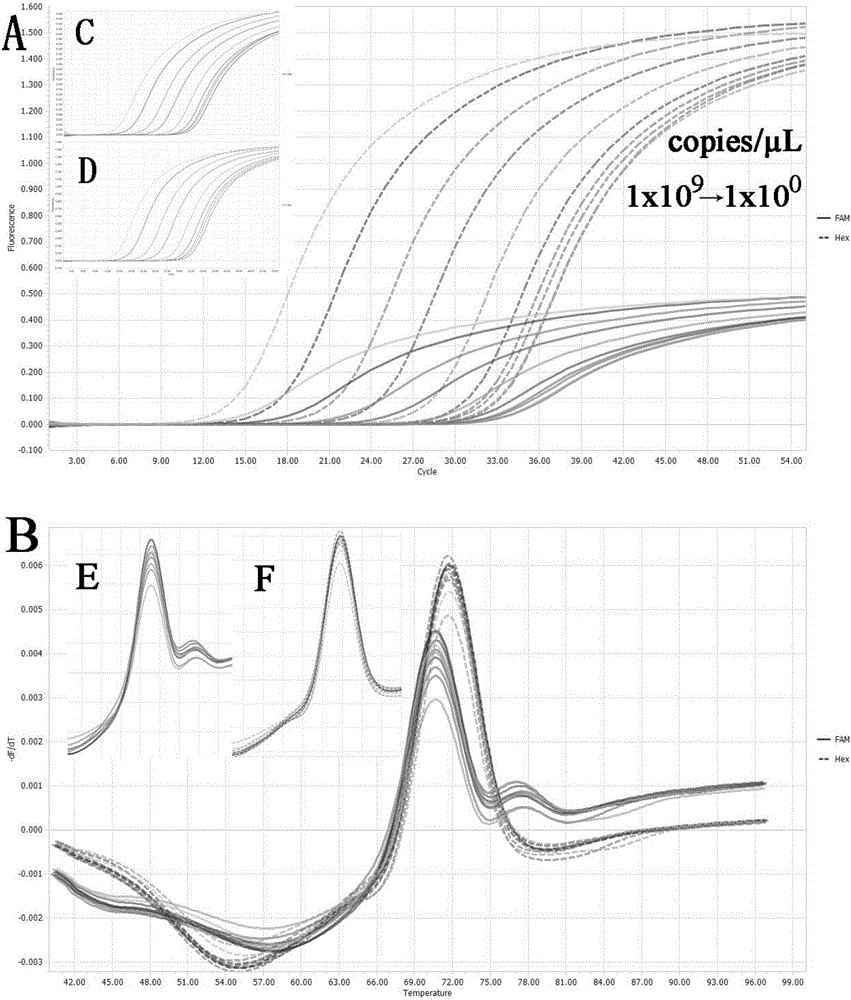
[0053] Example 2 Preparation of standard samples, two-color fluorescent PCR amplification and melting curve analysis
[0054] 1) Extraction of porcine pseudorabies virus DNA:
[0055] Samples of diseased materials suspected of being infected with PRV were taken respectively. The samples of diseased materials could be lymph nodes, brain, heart, liver, spleen, lung, kidney, tonsil and other tissues of dead pigs, or collected pig serum or nose samples. Serum can be taken directly in 200 μl for later use; the nose formula needs to be dissolved in 1mL PBS hydrochloric acid buffer solution, and 200 μl of it should be taken after standing for 10-20 minutes; Add 3mL PBS hydrochloric acid buffer solution to dissolve, take 200μL for later use. Nucleic acid extraction was carried out according to the instructions of TAKARA's MiniBEST Viral RNA / DNA Extraction Kit Ver.4.0.
[0056] 2) Preparation of standard samples:
[0057] In order to verify the feasibility and reliability of the met...
Embodiment 3
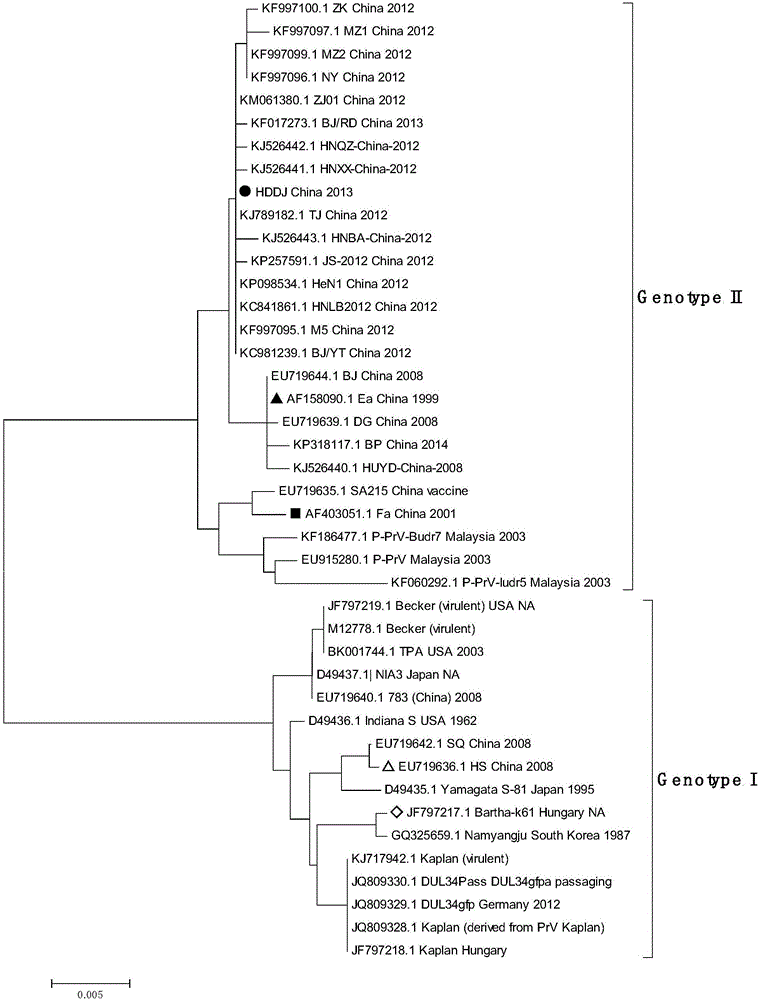
[0081] Embodiment 3 specificity experiment
[0082] Next, the detection method established by the present invention is used for specific detection.
[0083]Extract other common porcine reproductive disorders related viral nucleic acids, such as extraction of classical swine fever virus (classical swine fever virus, CSFV), porcine encephalitis virus (Japanese encephalitis virus, JEV), porcine circovirus type 2 (Porcine circovirus type 2 , PCV 2), porcine reproductive and respiratory syndrome virus (Porcine Reproductive and Respiratory Syndrome Virus, PRRSV), porcine parvovirus (Porcine parvovirus, PPV), the RNA of CSFV, JEV and PRRSV was reverse transcribed into cDNA, and the cDNA of the above virus , the nucleic acid and water of PCV 2 and PPV were used as PCR templates respectively, analyzed by the PCR amplification reaction and melting curve analysis method in (2) above, and tested with the porcine pseudorabies virus Bartha-K61 vaccine strain (also European and American type...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com