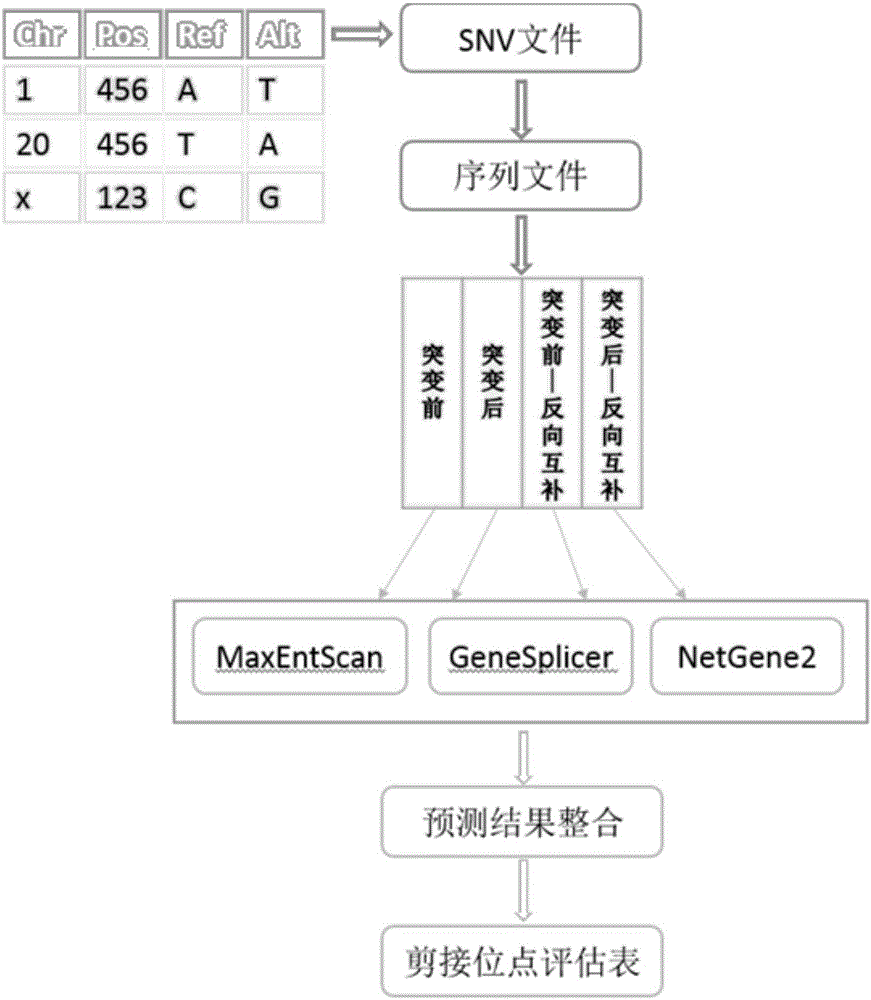
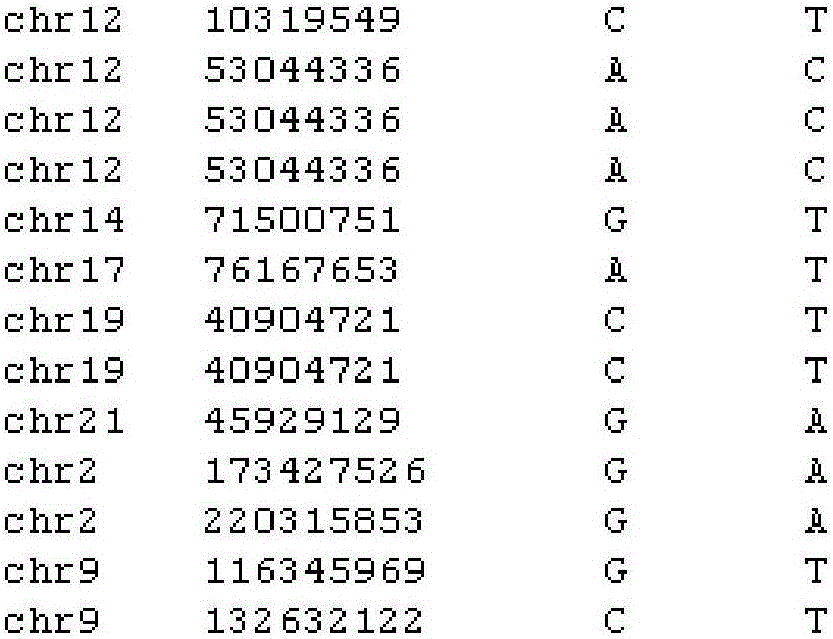
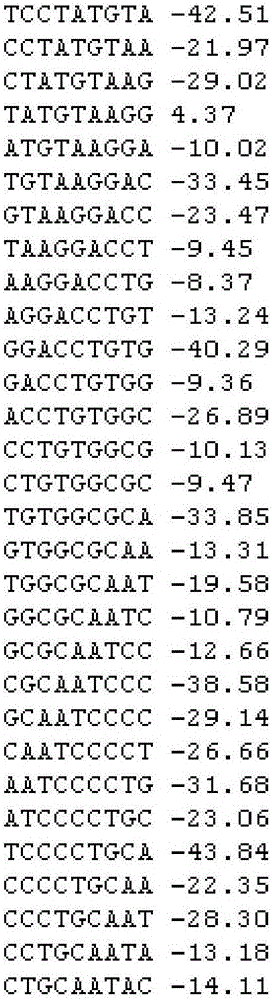SNV detection system affecting RNA splicing
An RNA splicing and detection system technology, applied in the field of SNV detection system, can solve the problems of RNA splicing change, affecting the effective translation of mRNA, etc., and achieve the effect of improving accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0027] It should be noted that, in the case of no conflict, the embodiments of the present invention and the features in the embodiments can be combined with each other.
[0028] The present invention will be described in detail below with reference to the accompanying drawings and examples.
[0029] Method principle of the present invention:
[0030] The present invention predicts the impact of SNV on RNA splicing, integrates three softwares based on different algorithms to predict RNA splicing sites, takes the SNV information file as input, and extracts the 100bp sequences upstream and downstream of the SNV site before and after the mutation and their directions The complementary sequence predicts the splicing site, thus, the change information of DNA positive strand and negative strand RNA splicing site before and after mutation is obtained.
[0031] First, the sequence is extracted. The present invention not only extracts the normal sequence fragments when no mutation occ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com