Method for detecting Psa in kiwi fruit pollens
A detection method and kiwifruit technology, applied in the directions of microorganism-based methods, microorganism determination/inspection, biochemical equipment and methods, etc., to achieve the effect of improving detection efficiency, enhancing specificity, and enhancing sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0035] Establishment of nested PCR detection method for kiwifruit Psa
[0036] (1) Cultivation of kiwifruit Psa and Pseudomonas syringae pv.tomato, Pseudomonas syringaepv.Theae, Pseudomonas avellanae: kiwifruit Psa was cultured with King’B liquid medium,
[0037] (2) DNA extraction of pathogenic bacteria: Ezup column genomic DNA extraction kit (bacteria) was used to extract target DNA.
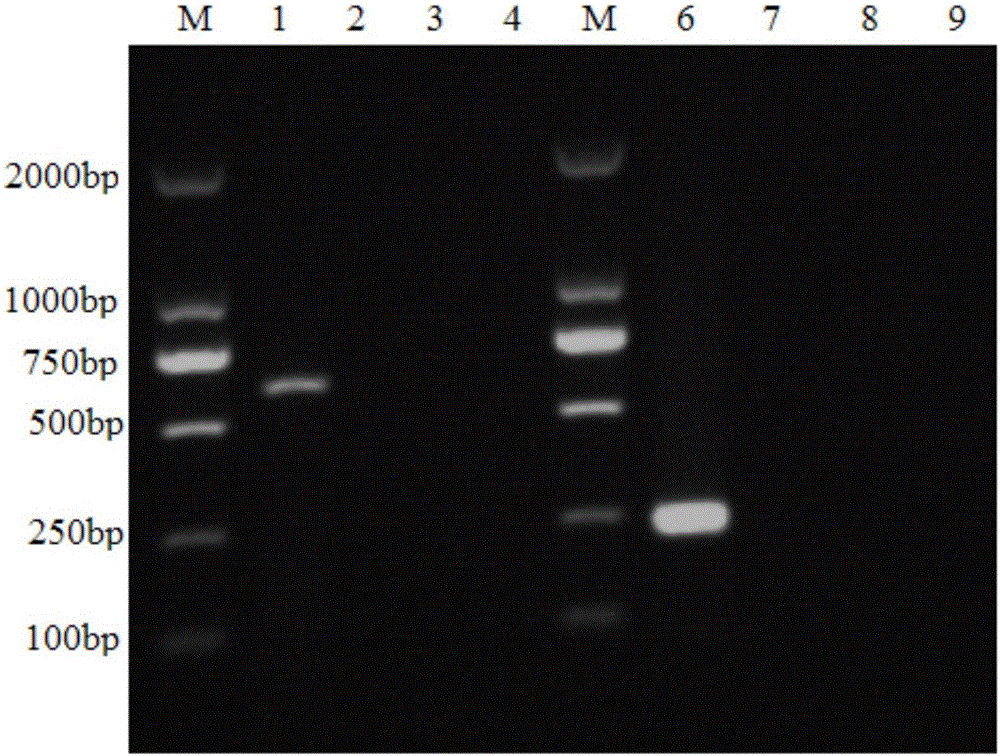
[0038] (3) Design of primers: two pairs of primers were designed according to the hrpw gene of Psa, the dominant pathogen of kiwifruit canker in Shaanxi.
[0039] The enrichment method of kiwi pollen Psa is as follows: Weigh 0.01 g of kiwi pollen, add it to a glass test tube filled with 15 mL of high-pressure sterilized and cooled King’s B liquid medium, seal it with a cotton plug, and place it in a constant temperature shaking incubator for 24 After culturing at 180r / min for 12 hours, store at 4°C until use.
[0040] The method of extracting the DNA of Psa in the pollen suspension is as fol...
Embodiment 2
[0050] Sensitivity of kiwifruit Psa PCR detection
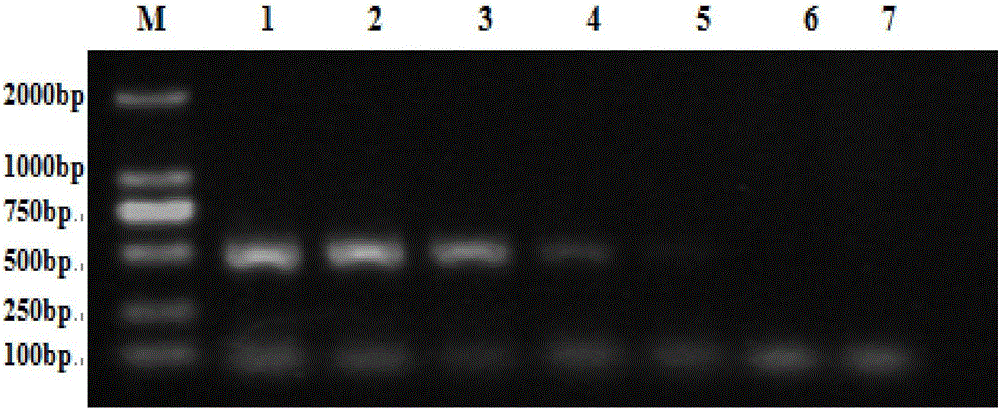
[0051] Pick a single colony of Psa and place it in 100 mL of sterile King’s B liquid medium, seal it with kraft paper, and culture it at 24°C and 200 r / min for 16 hours to obtain a bacterial suspension. Use a hemocytometer to measure the concentration of the bacterial suspension, and adjust the concentration gradient of the bacterial suspension to 10 with sterile normal saline. 7 cfu / mL, 10 6 cfu / mL, 10 5 cfu / mL, 10 4 cfu / mL, 10 3 cfu / mL, 10 2 cfu / mL, 10cfu / mL, extract DNA respectively, carry out PCR amplification, study the detection limit of Psa in the bacterial suspension, and determine the detection sensitivity of this method.
[0052] Depend on figure 2 It can be seen that: Swimming lanes 1-7: the concentration of the detected bacterial suspension is 10 in sequence 7 cfu / mL, 10 6 cfu / mL, 10 5 cfu / mL, 10 4 cfu / mL, 10 3 cfu / mL, 10 2 cfu / mL, 10cfu / mL, the target band becomes darker with the decrease of the conc...
Embodiment 3
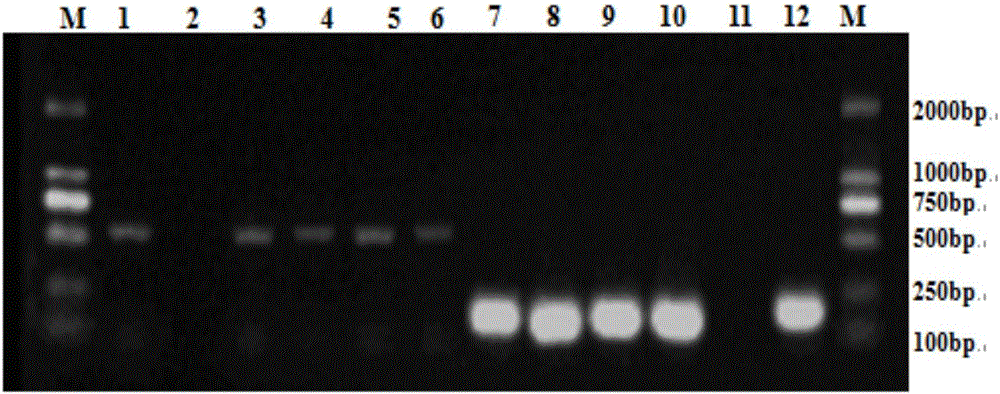
[0054] Detection of Psa PCR in kiwifruit pollen
[0055] (1) Cultivation of kiwifruit pollen Psa: Take 0.01 g of prepared kiwifruit pollen, respectively, and add them into glass test tubes numbered 1-23 containing 15 mL of autoclaved and cooled King’B liquid medium, and seal with cotton plugs. After culturing in a constant temperature shaking incubator at 24°C and 180r / min for 12h, a mixed bacterial suspension was obtained. Store at 4°C until use (short-term storage only).
[0056] (2) Extraction of Psa DNA from the bacterial suspension: Take 1 mL of the overnight cultured bacterial suspension in a 1.5 mL centrifuge tube, centrifuge at room temperature at 11,000 rpm for 1 min, discard the supernatant, and collect the bacterial cells. Add 1mLddH 2 O Pipette repeatedly, wash the cells, centrifuge at 11,000rpm for 1min, and discard the supernatant. Repeat wash once. Take 100 μL ddH 2 O, pipette repeatedly to dissolve the precipitate, put it in a water bath at 100°C for 15 mi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com