Sturgeon sexuality difference molecular marker and application thereof
A molecular marker, differential technology, applied in DNA/RNA fragmentation, microbial determination/inspection, recombinant DNA technology, etc., can solve the problems of individual size limitation, death of sturgeon, and high requirements for operating experience, and achieves convenient operation and method. Accurate and reliable, the effect of improving economic efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] Example 1 Obtaining Molecular Markers Related to Sturgeon Sex
[0027] 1.1 Acquisition of sexed sturgeon populations
[0028] The population used was Sturgeon's sturgeon with known sex from a sturgeon farm in Jiangsu, with 50 male and 50 fish each. The cut dorsal fin rays of the fish were stored in 95% ethanol at -20°C for genomic DNA extraction.
[0029] 1.2 Genomic DNA extraction from sturgeon
[0030] In this experiment, the conventional phenol-chloroform method was used to extract genomic DNA from sturgeon fin rays, and the specific steps were as follows:
[0031] (1) Take 0.3-0.5g of fin rays in a 1.5ml Eppendorf tube, cut it into pieces, and dry it for 20 minutes on a clean bench;
[0032] (2) After the ethanol is basically volatilized, wash with TE buffer solution (10mmol / ml Tris, 1mmol / ml EDTA, SDS 5%, pH=8.0) for 1-2 times, then add 600μl DNA extraction solution (0.001mol / LTris- C1, 0.1mol / L EDTA, SDS 5%, pH=8.0) and 3μl proteinase k (200mg / ml), digest in a...
Embodiment 2
[0037] Example 2 Verification of Molecular Markers Related to Sturgeon Sex
[0038] 2.1 Extraction of genomic DNA from fin rays of sturgeon to be verified
[0039] Sturgeons to be verified belong to the group in Example 1, 12 males and females were randomly selected, and the DNA extraction method was as described in Example 1.
[0040] 2.2 Amplification of nucleotide fragments containing male-specific reads
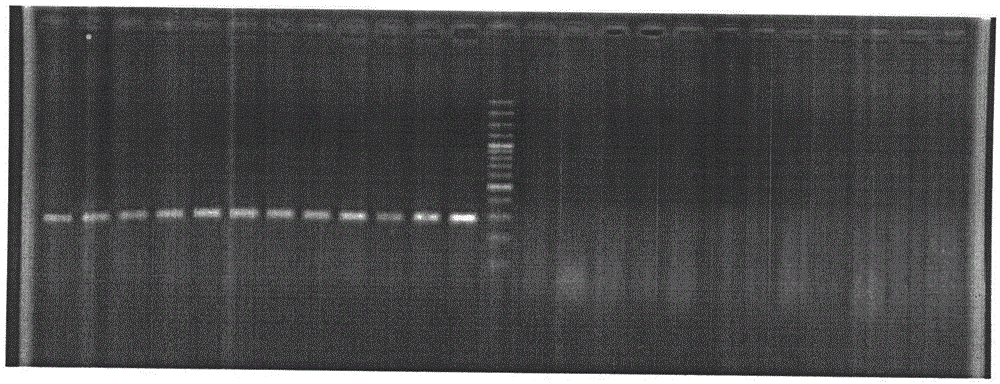
[0041] Primers were designed respectively according to the DNA sequences SEQ ID NO.4 and SEQ ID NO.5 of the male-specific reads, and the primer sequences are shown in SEQ ID NO.2 and SEQ ID NO.3. Using the genomic DNA in 2.1 as a template, the PCR reaction system in 25 μl is: 50-100ng / μl template DNA 1μl, 10pmol / μl primers F and R 1μl each, 10mmol / L dNTP mix 2.0μl, 5U / μl Taq DNA polymerase 0.125 μl, 2.5 μl of 10×PCR reaction buffer, and double-distilled water as the balance; PCR reaction conditions: 94°C for 5 minutes; 30 cycles of 94°C for 30 seconds, 55°C for 30 secon...
Embodiment 3
[0042] Example 3 Application of Molecular Markers Related to Sturgeon Sex
[0043] 3.1 Extraction of genomic DNA from the fin rays of the sturgeon to be tested
[0044] The sturgeons to be tested were Sturgeon's sturgeons of known sex from a sturgeon farm in Zhejiang, 12 males and 12 males were randomly selected, and the DNA extraction method was as described in Example 1.
[0045] 3.2 Amplification of sex-related molecular markers in sturgeon
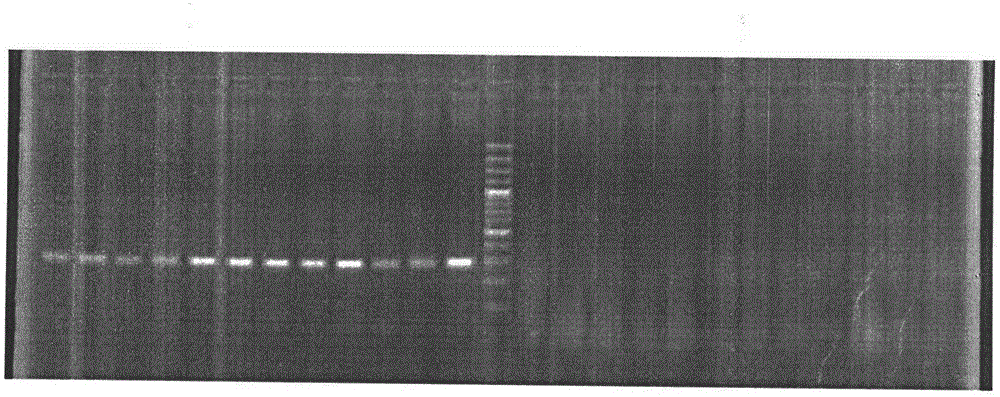
[0046] Use the primers whose sequences are shown in SEQ ID NO.2 and SEQ ID NO.3, use the genomic DNA in 3.1 as a template, and use the PCR reaction system and conditions in 2.2. The results showed that the amplified band of 286bp was male fish, while the female fish had no such amplified product. The agarose gel electrophoresis pattern of the amplified product is attached figure 2 As shown, the middle lane of the gel map is the molecular weight Marker, the uppermost band of the Marker is 1500bp, the lowermost band is 100bp, and the...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com