Nanofiber biological membrane immobilized bi-enzyme system and trehalose catalytic synthesis method thereof
A technology of nanofibers and biofilms, applied in the biological field, can solve the problems of low immobilization efficiency of single-enzyme catalytic systems, lack of competitive advantages in substrates, and low enzyme utilization efficiency, so as to improve immobilization efficiency and utilization efficiency, The effect of reducing the difficulty of separating and purifying trehalose and improving the conversion efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
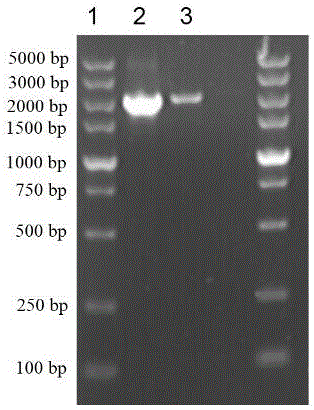
[0050] This example illustrates the construction of engineering bacteria co-expressing trehalose synthase gene TreS and amyloid CsgA-SpyTag fusion gene
[0051] The gene sequence of the fusion protein CsgA-SpyTag is based on the CsgA sequence published by NCBI (NCBI Reference Sequence: NC_000913.3) and the reported literature (Nguyen P Q, Botyanszki Z, Tay P K R, et al. Programmable biofilm-based materials from engineered curli In nanofibres[J]. Nature communications, 2014, 5.), SpyTag (polypeptide sequence AHIVMVDAYKPTK) is fused and connected by a flexible linker of 6 amino acid residues GSGGSG.
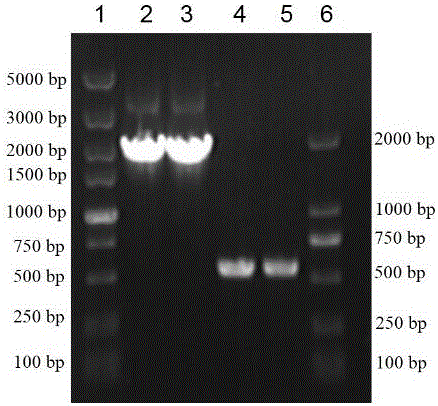
[0052] Specifically, the constructed strains E. coli BL21(DE3)(pET22b-treS)(Jiang L, Lin M, Zhang Y, et al. Identification and characterization of a novel trehalose synthase gene derived from saline-alkali soil metagenomes[J]. PloS one, 2013, 8(10 ): e77437.) inoculated into LB liquid medium (peptone 10 g / L, yeast powder 5 g / L, sodium chloride 10 g / L) at 37°C, 200 rpm for overn...
Embodiment 2
[0064] This example illustrates the construction of engineering bacteria expressing β-amylase-SpyCatcher fusion gene BA-SC.
[0065] The fusion protein BA-SpyCatcher gene sequence is based on the NCBI published β-amylase BA sequence (GenBank: AJ250858.1) and the published SpyCatcher gene sequence (GenBank: JQ478411.1) through the flexible linker fusion of 6 amino acid residues GSGGSG The BA sequence of β-amylase is obtained by whole gene synthesis method.
[0066] According to the β-amylase gene sequence and the SpyCatcher gene sequence, the upstream and downstream primers F1 and R1 (amplification of the β-amylase gene BA), F2 and R2 (amplification of the polypeptide tag SpyCatcher) were designed and synthesized.
[0067] F1:5'-CG GGATCC ATGTTCATTTTGAGTC-3'
[0068] R1: 5'-ACTGCCACCGCCACCGCTACCGCCACCGCCCCAATTTATCTGTATAA-3'
[0069] F2: 5'-ACTGCCACCGCCACCGCTACCGCCACCGCCCCAATTTATCTGTATAA-3'
[0070] R2: 5'-CC CTCGAG TTAAATATGAGCGTCACCTTTAGTT-3'
[0071] Among them, BamH ...
Embodiment 3
[0077] This example illustrates the effect of creating functional nanofibrous biofilms on the surface of trehalose synthase-containing cells
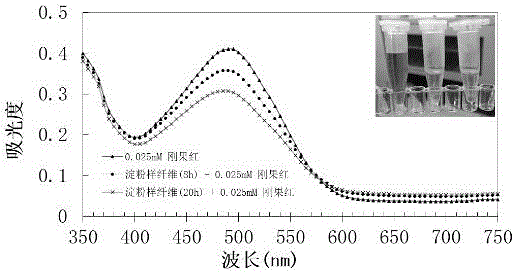
[0078] strains from the plate E. coli BL21(DE3) (PETDuet-TreS-CsgA-ST) was inoculated into 50ml LB medium containing ampicillin resistance, cultured overnight at 37°C, 200rpm / min for 12h; Resistance 50 mL autoinduction medium (glycerol 5g / L, Na2HPO4 6g / L, K2HPO4 3g / L, NH4Cl 1g / L, NaCl 0.5g / L, MgSO4-7H2O 0.5g / L, soybean peptone 10g / L ), 30°C, 180r / min shaking culture to induce expression for 10-12 h, collect the fermentation broth, wash twice with pH 7.0 PBS buffer and resuspend, the enzyme activity of trehalose synthase cells was determined to be 8000U / mL. The Escherichia coli containing trehalose synthase was bound and adsorbed by Congo red solution, and it was observed that Congo red was obviously adsorbed on the surface of the bacterial cells. At the same time, the Escherichia coli that could produce CsgA amyloid fibers was analyz...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com