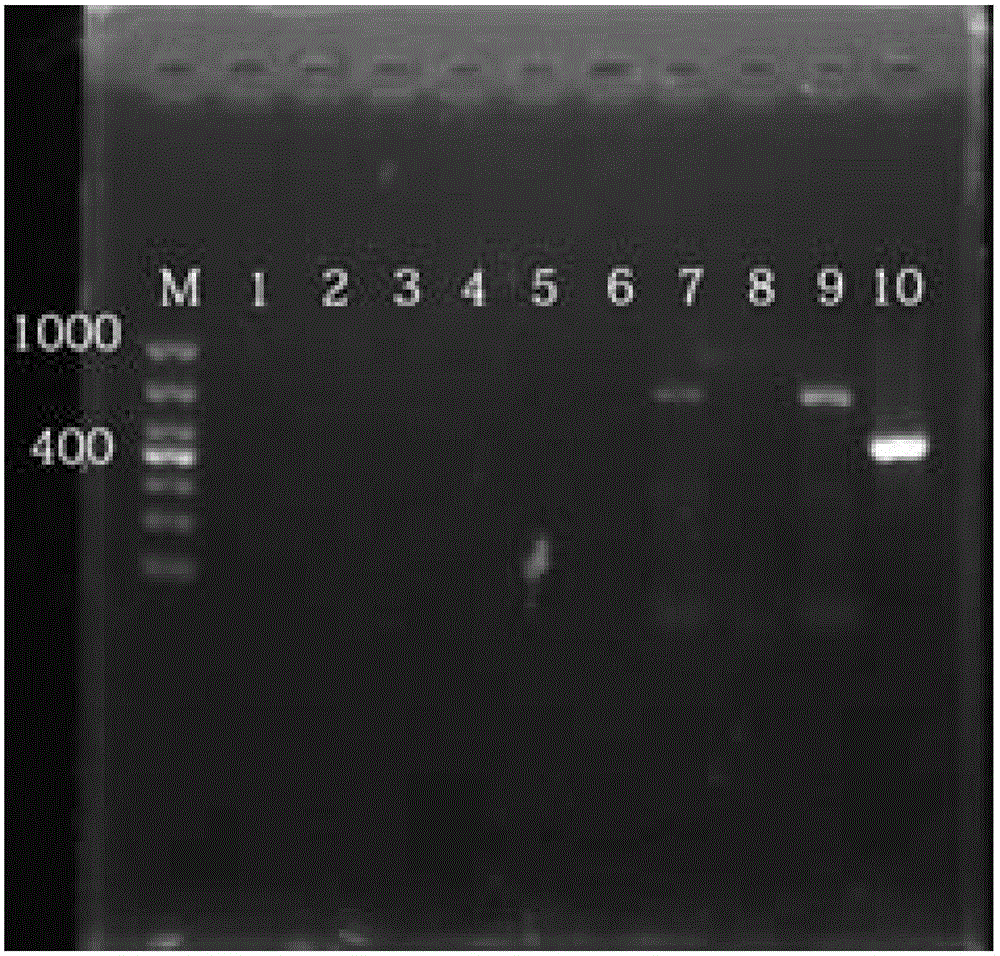
Nested RT-PCR detection method for differentiating variant strains and classic strains of PEDV
A technology of RT-PCR and detection method, which is applied in the field of rapid detection of PEDV virus, can solve the problems of declaration and authorization without the identification and detection technology of PEDV classic strain and variant strain.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] The extraction of RNA in the sample of embodiment 1
[0033] 1 Experimental materials
[0034] Samples containing PEDV strains were named MX-2014, MX-2015, HA-2015, NC-2014, NC-2015 according to the sample collection time and place; samples containing TGEV strains were named QW; samples containing PDCoV strains The samples containing the RV strain were designated as BC; and the samples containing the RV strain were designated as JC.
[0035] 1.1 Main reagents
[0036] Trizol total RNA extraction reagent; TaqTM enzyme, reverse transcriptase M-MLV, RNase inhibitor, 5×M-MLVbuffer, 10×PCR buffer, agarose, DL2000 DNAMaker, dNTPs, 6×Loadingbuffer were all purchased from Takara Company.
[0037] 2 test method
[0038] 2.1 Trizol method to extract total RNA
[0039] 2.1.1 Take samples MX-2014, MX-2015, HA-2015, NC-2014, NC-2015, QW, BC and JC grinding solution 250μL into 2mlEP tube, add 500μL pre-cooled Trizol.
[0040] 2.1.2 The samples were mixed vigorously and left at r...
Embodiment 2
[0045] Embodiment 2: Nested RT-PCR amplification reaction
[0046] 1 Design of primers
[0047] Two pairs of primers were designed and screened by Primer Premier 5.0 software. The upstream PEDV / F-1 of the outer primer: 5-AACACTTAGCCTACCACA—3', and the downstream PEDV / F-2 of the outer primer: 5'—GTGGAATCATTGGACAA—3'.
[0048] 2 The first nested RT-PCR amplification reaction
[0049] After total RNA was extracted by Trizol method, reverse transcription reaction was performed: 2 μL of RNA, 1 μL of PEDV / F-2 downstream primer and 3 μL of ddH2O were mixed, reacted at 70°C for 10 min, and then cooled on ice for two minutes.
[0050] 2.1 Add 2 μL 5×MLV buffer, 0.5 μL dNTPMixture, 0.25 μL RNase Inhibitor, 0.25 μL M-MLV and 1 μL ddH to this reaction mixture 2 O.
[0051] 2.2 After being incubated at 42°C for 1 hour, and then incubated at 70°C for 15 minutes, the reaction was terminated by cooling on ice to obtain cDNA.
[0052] 2.3 Take 2μL cDNA as a template for the first PCR ampli...
Embodiment 3
[0054] Embodiment 3: the second nested RT-PCR amplification reaction
[0055] 1 Design of primers for the second PCR
[0056] Two pairs of primers were designed and screened by Primer Premier 5.0 software. The upstream PEDV / R-1 of the internal primer: 5'-GAAAACCAGGGTGTCAA-3', and the downstream PEDV / R-2 of the internal primer: 5'-GAAATACCATCCTCCACCAG-3'.
[0057] 2 Carry out the second nested RT-PCR amplification reaction
[0058] Take 2 μL of the first PCR product as a template for the second PCR amplification, and configure a 25 μL reaction system: 10×PCR buffer 2.5 μL, dNTP Mixture 2 μL, external primers R1 and R2 1 μL each, r-Taq enzyme 0.5 μL, ddH 2 O16μL mix well.
[0059] 2.1 Amplify according to the following conditions: pre-denaturation at 94°C for 5 minutes; denaturation at 95°C for 5 minutes, renaturation at 51°C for 30 seconds, extension at 72°C for 30 seconds, 30 cycles; final extension at 72°C for 10 minutes to terminate the reaction. Obtain the second PCR pro...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com