Method for detecting whether Araliaceae plant components exist in sample and whether sample is adulterate
A technology for the detection of samples and plant components, applied in biochemical equipment and methods, determination/inspection of microorganisms, DNA/RNA fragments, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
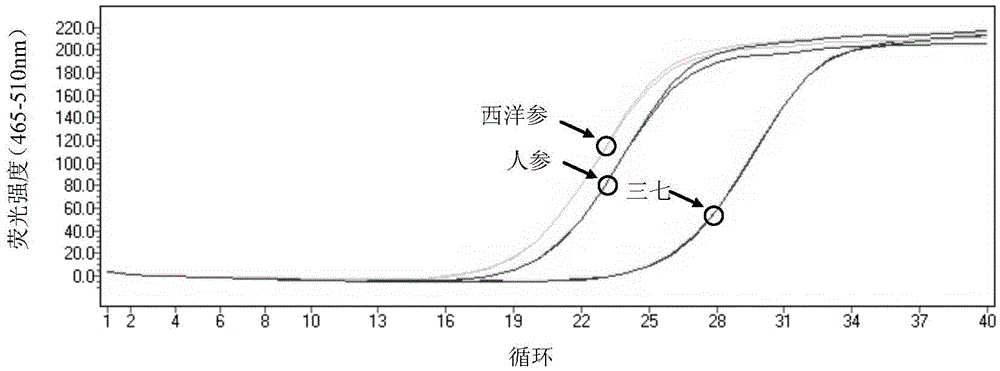
[0083] Embodiment 1SEQIDNO:1 and 2 to the PCR amplification of notoginseng, ginseng, American ginseng
[0084] (1) Take 30 mg of Panax notoginseng, Panax ginseng and Panax ginseng powder respectively, and extract Panax notoginseng, Panax ginseng, Total DNA of American ginseng, specifically:
[0085] (a) adding liquid nitrogen to fully grind;
[0086] (b) Quickly transfer the ground powder to a centrifuge tube pre-filled with 700 μL of 65°C preheated buffer GP1 (add mercaptoethanol to the preheated GP1 before the experiment to make the final concentration 0.1%), and quickly invert to mix After homogenization, place the centrifuge tube in a 65°C water bath for 20 minutes, and invert the centrifuge tube during the water bath to mix the sample several times;
[0087] (c) Add 700 μL of phenol / chloroform (v / v1:1), mix thoroughly, and centrifuge at 12000 rpm (~13400 × g) for 5 min;
[0088] (d) Carefully transfer the upper phase of the aqueous layer obtained in the previous step i...
Embodiment 2
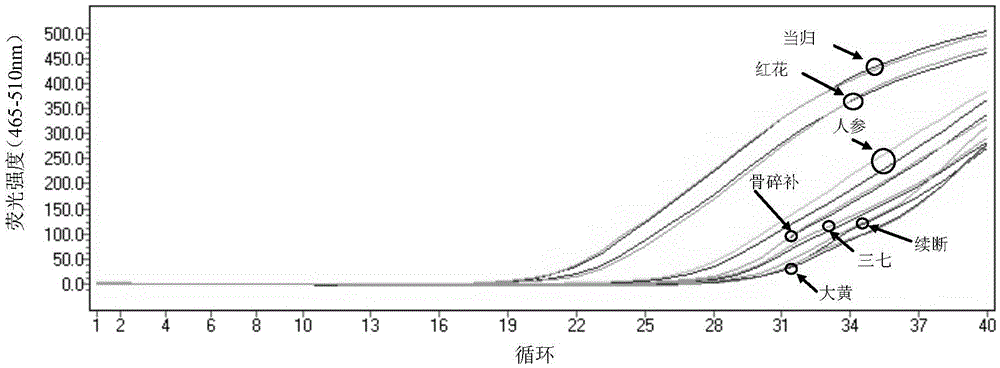
[0099] The comparison of embodiment 2SEQIDNO:1 and 2 and ITS2 universal primer
[0100] Taking the multi-flavored plant-derived medicinal materials in Dieda Huoxue Powder as an example, compare the PCR amplification results of SEQ ID NO: 1 and 2 with the ITS2 universal primer. The herbal medicinal materials in Dieda Huoxue Powder include safflower, angelica, rhizoma drynaria, Dipsacus, rhubarb and notoginseng medicinal materials, etc. Weigh 30 mg of each medicinal material powder and ginseng powder respectively, according to the step (1) in Example 1 Under the same conditions, extract the total DNA of each medicinal material; for the total DNA of each medicinal material, use the ITS2 universal primer and SEQ ID NO: 1 and 2 respectively, according to the step (2) in Example 1, carry out under the same conditions Real-time fluorescent PCR amplification, wherein, each sample is tested twice, and the sequence of the ITS2 universal primer is:
[0101] ITS2-3R: GACGCTTTCTCCAGACTACA...
Embodiment 3
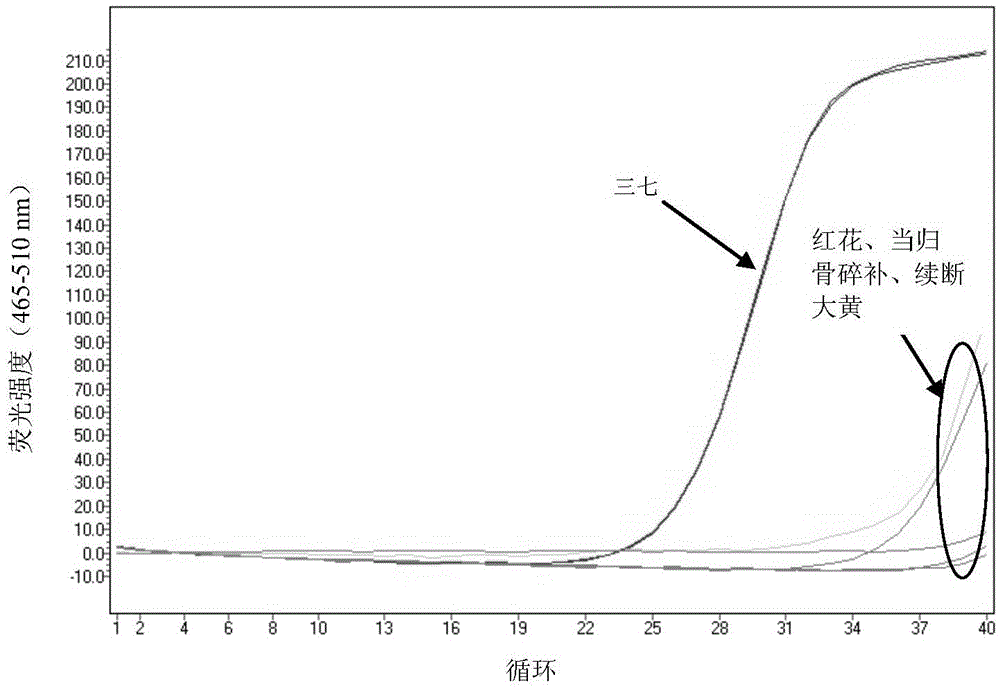
[0106] Embodiment 3 Utilize SEQIDNO:1 and 2 to identify whether adulteration in the notoginseng sample to be detected
[0107] Take by weighing the very fine powder of 30mg Radix Notoginseng, Radix Ginseng, American Ginseng respectively, by the step (1) in the embodiment 1, extract the total DNA of Radix Notoginseng, Radix Radix Radix Ginseng, American Ginseng respectively under the same conditions; Using SEQIDNO: 1 and 2 as primers, according to the step (2) in Example 1, carry out real-time fluorescent quantitative amplification under the same conditions, and test the high-resolution melting curve of the amplification result obtained, the test condition is 95 ° C denaturation , renatured at 40°C for 30s, kept at 65°C for 1s, slowly increased the temperature from 65°C to 95°C at a rate of 4.4°C / s until the DNA double strands were completely opened, collected fluorescence during the heating process, and tested each sample twice. The results are as follows Figure 5A and Figu...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com